Guohui Hu1#, Shiwen Luo1, Hai Rao2, Haili Cheng2*# and Xin Gan3#
1Center for Experimental Medicine, the First Affiliated Hospital, Nanchang University, Nanchang, P.R. China
2Department of Molecular Medicine, the University of Texas Health, San Antonio, TX 78229, USA
3Research Institute of Respiratory Medicine, the First Affiliated Hospital, Nanchang University, Nanchang, P.R China
These authors contributed equally to this work.
Corresponding Author:
Haili Cheng
Department of Molecular Medicine
the University of Texas Health
7703 Floyd Curl Dr., San Antonio, TX 78229, USA
Tel: 210-562-4149
Fax: 210-562-4161
E-mail: chengh3@livemail.uthscsa.edu
Received date: February 16, 2018; Accepted date: March 05, 2018; Published date: March 07, 2018
Citation: Hu G, Luo S, Rao H, Cheng H, Gan X (2018) A Simple PCR-based Strategy for the Introduction of Point Mutations in the Yeast Saccharomyces cerevisiae via CRISPR/Cas9. Biochem Mol Biol J Vol. 4: No. 1:9. doi:10.21767/2471-8084.100058
Keywords
Yeast; Mutagenesis; Integration; CRISPR; Genome editing
Introduction
The budding yeast Saccharomyces cerevisiae serves as a powerful model system with its facile genetics, wellcharacterized genomics, and a vast array of biochemical assays and tools [1-4]. Yeast research has provided key insights into the mechanisms underlying fundamental cellular processes (e.g., gene expression, DNA repair and proteolysis) and facilitated the development of numerous techniques including DNA transformation, genome manipulation and synthetic biology [1,3,4]. Nevertheless, existing approaches for the incorporation of specific mutations in the yeast genome remain tedious, laborious and inefficient [5-7] as they often require specialized systems (e.g., plasmids or conditions) and/or manipulating selectable markers, which introduces additional sequences that may have unintended effects [6,7].
The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system is emerging as a powerful geneediting tool in various organisms from yeast to human [8-10]. The RNA-guided endonuclease Cas9 can be targeted to specific genomic locus to generate a double-strand break, which allows the introduction of precise mutations by providing a DNA template for homologous repair. With its high efficacy and marker-free exercise, the CRISPR-Cas9 system overcomes many existing limitations and is becoming a preferred method for in vivo genome manipulation [9,11-13]. Among several yeast CRISPR approaches [11-13] derived from the work of DiCarlo et al. which first demonstrated the utility of CRISPR in the yeast [9], the protocol developed by Laughery et al. stood out with its higher efficiency and relative simplicity [11]. Nevertheless, one time-consuming step in Laughery’s approach involves the cloning of the single-guide RNA (sgRNA), which target Cas9 to relevant chromosomal loci [11], into the vectors (e.g., pML104, pML107) with unique restriction sites (SwaI and BclI). The strategy developed appeared to be faster, more convenient and efficient than several other CRISPR approaches in yeast [9,11-15], but still requires ~3-4 days to obtain the plasmids with right guide sequences (Figure 1) [11]. As previously described, the vector plasmids need to be transformed first into a dam- E. coli strain so that the methylation free plasmid can be digested by BclI; and overnight incubation is suggested for subsequent SwaI digestion; then the linearized plasmid is purified after gel electrophoresis and ligated overnight with oligonucleotides containing guide sequences (Figure 1) [11]. Then the plasmids constructed can be introduced to yeast for efficient genome editing [9,11].
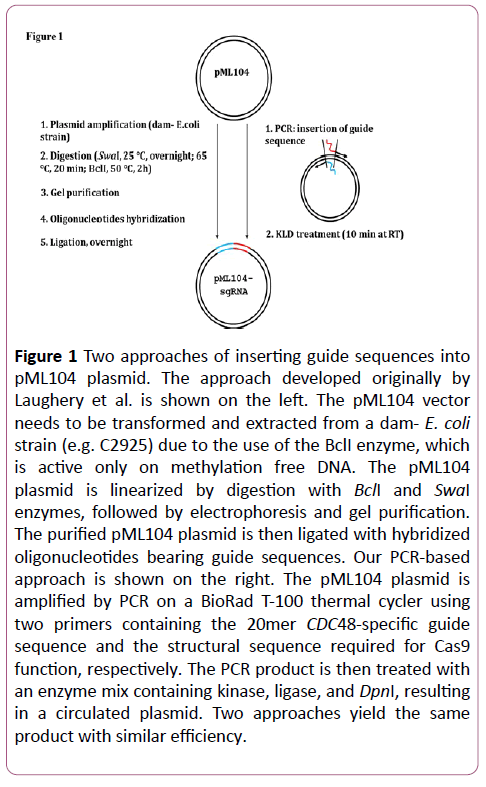
Figure 1: Two approaches of inserting guide sequences into pML104 plasmid. The approach developed originally by Laughery et al. is shown on the left. The pML104 vector needs to be transformed and extracted from a dam- E. coli strain (e.g. C2925) due to the use of the BclI enzyme, which is active only on methylation free DNA. The pML104 plasmid is linearized by digestion with BclI and SwaI enzymes, followed by electrophoresis and gel purification.The purified pML104 plasmid is then ligated with hybridized oligonucleotides bearing guide sequences. Our PCR-based approach is shown on the right. The pML104 plasmid is amplified by PCR on a BioRad T-100 thermal cycler using two primers containing the 20mer CDC48-specific guide sequence and the structural sequence required for Cas9 function, respectively. The PCR product is then treated with an enzyme mix containing kinase, ligase, and DpnI, resulting in a circulated plasmid. Two approaches yield the same product with similar efficiency.
Materials and Methods
Yeast cultures were grown in rich (YPD) or synthetic media containing standard ingredients. The pML104 derived plasmids were transformed to yeast strain BY4741 (Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) by standard LiAc based protocol [1,11]. The transformants were spread onto synthetic plates lacking uracil and grown for 3 days at 30°C. Colonies were inoculated in 3 ml YPD medium and grown overnight. The genomic DNA was extracted for PCR amplification of the region surrounding CDC48, which was then analyzed by sequencing to ascertain the incorporation of R105G mutation. Oligonucleotides were obtained from the Integrated DNA Technologies (Skokie, IL). PCR conditions are detailed below.
Results and Discussion
To generate Cdc48 R105G mutant, two primers were designed for inserting relevant guide sequences to target Cas9 specifically to the region around CDC48 R105. Primer MutpML- F (i.e. 5’- GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC-3’) contains the 20 nucleotides (unlined) of the structural element required for Cas9 function (Figures 2A and 2B). Primer Mut-pMLC48R105G- R2 (i.e. 5’- GCCTAATACGTAAATTGTTAGATCATTTATCTTTCACTGC-3’) bears the 20mer guide sequence of CDC48 (Figures 2A and 2B). These two oligonucleotides were used to PCR-amplify the plasmid pML104 using the Q5 site-directed mutagenesis protocol (New England Biolabs) with minor modifications. Specifically, in a 25 l PCR reaction, 8 ng of pML104 plasmid was mixed with 15 M of each primer and 12.5 l of 2x Q5 master mix. The PCR reaction was performed as follows: an initial denaturation step at 98 for 1 min, followed by 25 cycles of denaturation at 98 for 10 s, annealing at 55 for 30s, and extension at 72 for 8 min; a final extension step of 5 min at 72. PCR products were resolved by gel electrophoresis to confirm the amplification (Figures 2C and 2D).
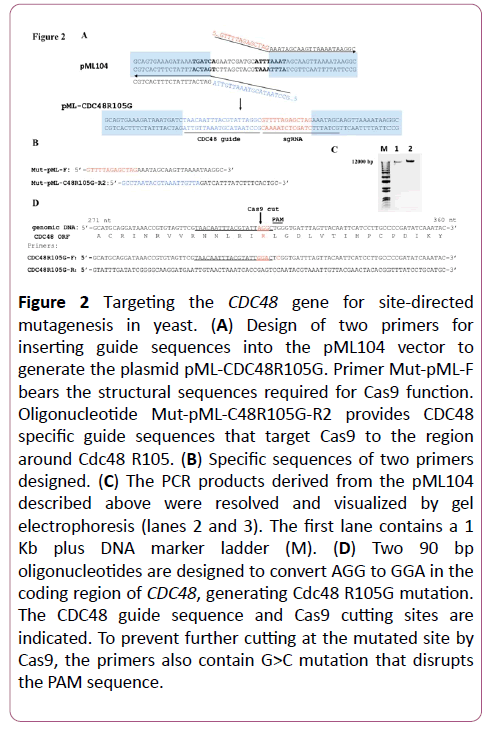
Figure 2: Targeting the CDC48 gene for site-directed mutagenesis in yeast. (A) Design of two primers for inserting guide sequences into the pML104 vector to generate the plasmid pML-CDC48R105G. Primer Mut-pML-F bears the structural sequences required for Cas9 function. Oligonucleotide Mut-pML-C48R105G-R2 provides CDC48 specific guide sequences that target Cas9 to the region around Cdc48 R105. (B) Specific sequences of two primers designed. (C) The PCR products derived from the pML104 described above were resolved and visualized by gel electrophoresis (lanes 2 and 3). The first lane contains a 1 Kb plus DNA marker ladder (M). (D) Two 90 bp oligonucleotides are designed to convert AGG to GGA in the coding region of CDC48, generating Cdc48 R105G mutation. The CDC48 guide sequence and Cas9 cutting sites are indicated. To prevent further cutting at the mutated site by Cas9, the primers also contain G>C mutation that disrupts the PAM sequence.
The PCR products were then incubated with the mixture containing kinase, ligase and DpnI enzymes provided in the Q5 site-directed mutagenesis kit (Figure 1), which promotes rapid circularization of the PCR products and removal of the template DNA. The reaction was performed at room temperature for 10 min and contained 2 l PCR products, 5 l 2x KLD reaction buffer, 1 l KLD enzyme mix, and 2 l nuclease-free water. Then it was transformed into competent E. coli cells C2987. Plasmid DNA was extracted from the resulting colonies and sequenced by T3 primer. Typically >60% colonies contained right inserts.
The resulting plasmid pML104-CDC48R105G could bring the Cas9 nuclease to the region surrounding R105 of Cdc48 to make a cut (Figure 2D). To generate Cdc48 R105G substitution, two 90 bp oligonucleotides were designed to replace AGG with GGA, and the protospacer-adjacent motif (PAM) was also disrupted by replacing TGG with TCG without changing amino acid (Figure 2D), which prevents subsequent Cas9 cutting in newly generated mutant cells. The plasmid pML104-CDC48R105G and hybridized double-stranded oligonucleotides CDC48R105G-F/CDC48R105G-R were co-transformed into yeast strain BY4741 and selected on plates containing synthetic complete media lacking uracil (SD-URA) [11]. Genomic DNA was then extracted from the resulting colonies and sequenced. Over 50% of colonies harbored CDC48 R105G mutation.
Conclusion
Here we have demonstrated an efficient PCR-based procedure for precise genome editing of yeast cells. This revised method retains high efficiency but eliminates the need of the dam- strain and markedly shortens the elaborate multistep cloning process, which includes overnight restriction digest and ligation, in creating the sgRNA targeting plasmid [11]. To incorporate sgRNA sequences to the pML104 plasmid, it takes ~5 hours with our modified method instead of ~3-4 days with the approach originally developed by Laughery et al. (Figure 1) [11]. The modifications introduced lead to significant improvements in substantial time, labor and cost savings. Our approach can be easily adapted for other organisms and may facilitate high-throughput generation of CRISPR-based genome engineering.
Acknowledgements
H. R. is supported by grants from the National Institutes of Health (GM 118350), the William & Ella Owens Medical Research Foundation, the Cancer Prevention Research Institute of Texas (RP170686), San Antonio Cancer Council and the National Center for Advancing Translational Science (UL1TR001120).
Author Contributions
G.H., H.C. and X.G. carried out the experiments. All authors analyzed the data. H.C. conceived and designed the experiments with inputs from H.R. H.C. and H.R. wrote the paper with the input from all authors.
Competing Interests Statement
The authors declare no competing interests.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, et al. (1996) Current protocols in molecular biology. New York: Wiley-Interscience, USA.
- Ju S, Tardiff DF, Han H, Divya K, Zhong Q, et al. (2011) A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol 9: e1001052.
- Vidal M, Fields S (2014) The yeast two-hybrid assay: still finding connections after 25 years. Nat Methods 11: 1203-1206.
- Usaj M, Tan Y, Wang W, VanderSluis B, Zou A, et al. (2017) TheCellMap.org: A web-accessible database for visualizing and mining the global yeast genetic interaction network. G3 (Bethesda) 7: 1539-1549.
- Storici F, Lewis LK, Resnick MA (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol 19: 773-776.
- Stuckey S, Mukherjee K, Storici F (2011) In vivo site-specific mutagenesis and gene collage using the delitto perfetto system in yeast Saccharomyces cerevisiae. Methods Mol Biol 745: 173-191.
- Finney-Manchester SP, Maheshri N (2013) Harnessing mutagenic homologous recombination for targeted mutagenesis in vivo by TaGTEAM. Nucleic Acids Res. 41: e99.
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60: 174-182.
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, et al. (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41: 4336-4343.
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, et al. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 154: 442-451.
- Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, et al. (2015) New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32: 711-720.
- Bao Z, Xiao H, Liang J, Zhang L, Xiong X, et al. (2015) Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol 4: 585-594.
- Horwitz AA, Walter JM, Schubert MG, Kung SH, Hawkins K, et al. (2015) Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas9. Cell Syst 1: 88-96.
- Jakociunas T, Bonde I, Herrgard M, Harrison SJ, Kristensen M, et al. (2015) Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 28: 213-222..
- Baek GH, Cheng H, Choe V, Bao X, Shao J, et al. (2013) Cdc48: a swiss army knife of cell biology. J Amino Acids 183421.



