Keywords
Alfalfa; Rubisco purification; Hydrolysis process; Proteinase-K; Rubisco
modelling; Biotechnological tools
Introduction
Proteins are widespread worldwide, they are indispensable for
humans and animals as they are essential for covering amino acid
requirements [1,2]. Animal meat is known as the primary source
of protein for human nutrition. However, following the strong
global population growth, several studies and investigations
have shown that meeting the need for animal protein seems
particularly difficult in the future [3]. Therefore, it is necessary
to double the meat production [4] but this solution will cause an
increased demand for animal feed as it consumes approximately
¾ of the world's produced biomass [5] and subsequently, heavily
affects the availability of vegetation and arable soil which are
limited. For this reason, we are looking for new sources of
protein to replace those that are no longer effective and which are cheaper, healthier, more nutritious and friendlier for the
environment [6]. Pulses and cereals are the non-animal source
for humans [7] and in recent years, plant leaves are considered to
be the main source of vegetable protein [8-10]. Alfalfa (Médicago
sativa L.) ranks among the most important vegetable protein
sources in the world. It is a forage legume grown on 33 million
hectares in the whole world [11]. It is known for its high forage
quality, its contribution in nitrogen fertilization and its high
protein content (enrichment of animal feed). Thus, in addition to
the use in the feeding of cattle, alfalfa enters into human nutrition
like sprouted seeds and juices; this is due to its valuable protein
considering alfalfa as a source of secondary metabolites for the
agricultural and pharmaceutical industry [12]. This legume was characterized by its protein-rich leaves and in particular Rubisco
that is the most abundant enzyme in our planet [13] and it
constitutes about 65% of the soluble alfalfa leaf protein [10]. This
intake, essential amino acids for human health, mineral salts and
macro and micro nutrients [14]. Also, its antioxidant capability
and its richness in phytoestrogens, saponins and vitamins allow
bioactive molecule has been extracted from different plant parts
(roots, stems, flowers and leaves) [15,16] and it is considered to
be a protein complex having a molecular weight of 560 kDa and
consisting of eight copies of large protein chain (55 kDa) [17] and
eight copies of a small protein chain (14 kDa) [18].
It is crucial and so important to signal that Rubisco seems to be a
new potential source of bioactive peptides [16]. Indeed, the goal
of this work is to extract Rubisco from Gabesian alfalfa leaves
(southern Tunisia) that was the most salt tolerant one among 12
accessions lines studied by our group (data in publication) and
there were different methods allow the extraction of concentrated
total proteins [19,20]. In our study we followed methodology
described by Kobbi S, et al. [10] for the Rubisco extraction and
purification from Alfalfa leaves. Further, bioactive peptides from
extracted Rubisco were done using hydrolysis by proteinase-K.
This methodology reduces complexity, cost and environmental
problems associated with conventional industrial processes.
Materials and Methods
Biological materials
50g of Gabesian alfalfa leaves (the region of Limaoua) were
used. The leaves were harvested, crushed and cold pressed.
The resulting paste was dried in a furnace at (37°C) to obtain an
heterogeneous dried alfalfa powder. Then, the last one was ready
for Rubisco purification. This method is the dehydration process
as described by Incamps et al. [21].
Extraction and purification of Rubisco
After alfalfa leaves dehydration process, 10 g of powder with
high protein content was obtained and the protein extraction and purification were carried out as indicated in [10]. In order
to isolate the soluble fraction, 10g of the alfalfa powder was
solubilized in 100 mL of distilled water at pH 10 by the addition
of NaOH (0.1 M). A first centrifugation was made at 10000xg for
20min at 4°C to eliminate the green protein and all debris. The
supernatant containing Rubisco was recovered and the pellet was
centrifuged again to retrieve the second supernatant. After four
successive centrifugations at pH 10, four solutions were summed
up to recover all Rubisco. Then, proteins were precipitated by
the acidification of the mixture at pH 3 using 10 mM of chloridric
acid (37% PA-ACS-ISO, parchased from Panreac) to precipitate the
proteins. This was followed by a last centrifugation at 10000xg
for 20 min at 4°C. The precipitate was recovered and the Rubisco
was then concentrated by glacial acetone (analytical grade, ACS,
Reag. Ph Eur, purchased from Scharlau under matricule number
of AC03111000).
Protein assay and electrophoretic mobility
The protein content was determined using the Biorad-Bradford
assay with absorbency reading at 595nm. The Bovine Serum
Albumin (BSA) was used as protein standard [22]. Polyacrylamide
gel electrophoresis in the presence of sodium dodecyl sulfate
(SDS-PAGE) was carried out in 15% polyacrylamide gel following
the standard procedure by Laemmli and Favre [23].
Optimization of Rubisco hydrolysis
The hydrolysis was carried out by mixing 1mL of the purified
Rubisco with proteinase-K. Then the solution was hydrolyzed in a
water bath [24] and the reaction was stopped by ice. The influence
of temperature, time of incubation and also proteinase-K
concentration was studied using the ‘one variable at a time’
method. The Box-Behnken Response Surface Methodology was
applied to optimize the three factor levels for increasing the
efficiency of peptides production. These three independent
parameters were taken at three levels (–1, 0, +1) and 15 assays
were performed as demonstrated in Table 1.
| Run No. |
[Protease-K] (mg/mL) |
Time(h) |
Temperature (°C) |
Yields (%) |
| Actual value |
Coded value |
Actual value |
Coded value |
Actual value |
Coded value |
| 1 |
6 |
(-1) |
6 |
(-1) |
43 |
(0) |
60.55 ± 0.11 |
| 2 |
6 |
(-1) |
10 |
(+1) |
43 |
(0) |
69.15 ± 0.25 |
| 3 |
20 |
(+1) |
6 |
(-1) |
43 |
(0) |
72.75 ± 0.19 |
| 4 |
20 |
(+1) |
10 |
(+1) |
43 |
(0) |
67.22 ± 0.15 |
| 5 |
13 |
(0) |
6 |
(-1) |
30 |
(-1) |
72.97 ± 0.23 |
| 6 |
13 |
(0) |
6 |
(-1) |
56 |
(+1) |
54.35 ± 0.12 |
| 7 |
13 |
(0) |
10 |
(+1) |
30 |
(-1) |
74.62 ± 0.18 |
| 8 |
13 |
(0) |
10 |
(+1) |
56 |
(+1) |
60.40 ± 0.21 |
| 9 |
6 |
(-1) |
8 |
(0) |
30 |
(-1) |
71.32 ± 0.27 |
| 10 |
20 |
(+1) |
8 |
(0) |
30 |
(-1) |
70.25 ± 0.19 |
| 11 |
6 |
(-1) |
8 |
(0) |
56 |
(+1) |
55.75 ± 0.14 |
| 12 |
20 |
(+1) |
8 |
(0) |
56 |
(+1) |
71.12 ± 0.24 |
| 13 |
13 |
(0) |
8 |
(0) |
43 |
(0) |
64.00 ± 0.22 |
| 14 |
13 |
(0) |
8 |
(0) |
43 |
(0) |
64.15 ± 0.29 |
| 15 |
13 |
(0) |
8 |
(0) |
43 |
(0) |
64.97 ± 0.27 |
Table 1: The matrix illustrating the conditions that were used to optimize the hydrolysis of the Rubisco by protease-K.
This hydrolysis was done at different temperatures (30, 43, and
56°C), at different enzyme concentrations (6, 13, and 20 mg/mL)
and at different times (6, 8, and 10 h) to investigate the kinetics
hydrolysis of Rubisco.
The mean values (n=3) of the yield were considered as responses.
The yield was fitted according to the second-order polynomial
model:

In this equation, Y is the production yield, a0 is the intercept
value, ai is the linear value, aii is the second order value, aij is
the interaction term and Xi is the independent parameters. The
response surface diagrams were shown using the SPSS software
(IBM SPSS version 21).
DPPH radical-scavenging assay
DPPH radical-scavenging activity of the alfalfa crude was
determined as described by Bersuder et al. and Hsouna et al.
[25,26]. To do this, 500 μL of the sample was mixed with 500 μL
of 99.5% ethanol and 125 μL of 0.02% DPPH in 99.5% ethanol.
Then the mixture was kept in the dark room for 60 min, and the
reduction of DPPH radical was measured at 517 nm. The control
was conducted in the same manner; except that distilled water
was used instead of sample and Butylate-dhydroxy-toluene (BHT)
was used as standard. The DPPH activity was calculated as follows:
(%) = [1-(Absorbance of sample/Absorbance of negative control)]
× 100.
Statistical analysis
All data were submitted to Analysis of Variance (ANOVA) and
differences between means were evaluated by Duncan’s Multiple
Range Test. The data were analyzed using the Statistical Package
for the Social Sciences (SPSS) version 10.0 (Chicago, Illinois, USA).
Differences were considered significant at p <0.05
Results and Discussion
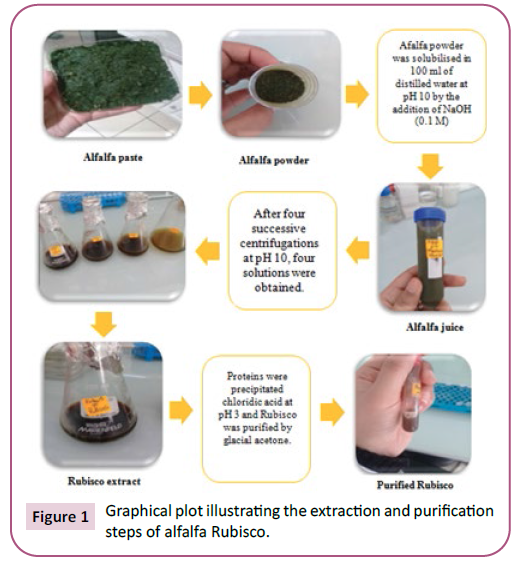
Production and purification of Rubisco
Rubisco is a bioactive molecule that has been labeled the most
abundant and largest protein on earth [10,27]. In our study, the
dried green powder was prepared from alfalfa leaves in order
to extract and purified Rubisco at homogeneity. Indeed, and as
indicated in Figure 1, after three centrifugation of alfalfa green
juice, we recovered the major Rubisco fraction. Then, the Rubisco
solution was centrifuged and the recuperated supernatant was
precipitated using the acetone concentration protocol. Proteins
content of the purified Rubisco was estimated to 40 mg/mL.
Figure 1: Graphical plot illustrating the extraction and purification
steps of alfalfa Rubisco.
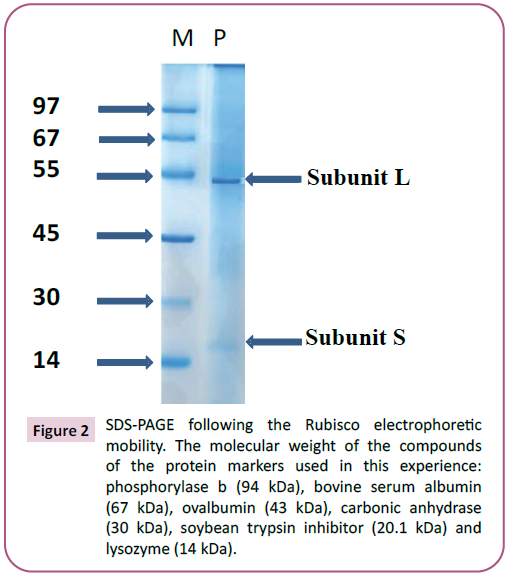
SDS-PAGE and in silico analysis of the Rubisco
The Rubisco obtained from the green juice was analyzed by
SDS-PAGE on 15% (w/v) to confirm their purity. Indeed, Figure 2 confirms that the studied protein was purified at homogeneity
and is composed of large subunit (L) possessing an apparent molecular weight of 55 kDa and small subunit (S) possessing
an apparent molecular weight of 13 kDa. Quaternary structural
studies have previously demonstrated that native Rubisco is a
protein consisting of eight small and eight large subunits [28,29].
Figure 2: SDS-PAGE following the Rubisco electrophoretic
mobility. The molecular weight of the compounds
of the protein markers used in this experience:
phosphorylase b (94 kDa), bovine serum albumin
(67 kDa), ovalbumin (43 kDa), carbonic anhydrase
(30 kDa), soybean trypsin inhibitor (20.1 kDa) and
lysozyme (14 kDa).
On the same case, Table 2 shows that Rubisco is composed of
569 amino acids: 446 amino acids in the large subunit whish their
molecular weight is about 55 kDa and 123 amino acids in the
small subunit whish their molecular weight is about 13 kDa. The
theoretical isoelectric point is computed to be 6.6 to 6.9 for the
subunit L and 7.45 for the subunit S (Table 2).
| Rubisco characteristics |
Large subunit |
Small subunit |
| Amino acid composition |
446 |
123 |
| Molecular weight |
55 kDa |
14 kDa |
| Theoretical isoelectric point |
6.6 to 6.9 |
7.45 |
Table 2: Illustration of some of the Rubisco in silico parameters such as the
amino acid composition, the molecular weight and also the theoretical
isoelectric point.
Optimization of the proteolytic Rubisco
hydrolysis
The Box-Behnken Response Surface Methodology was monitored
in order to determine the effects of the three selected variables
(the reaction temperature (°C); the reaction incubation time
(h) and the proteinase-K concentration (mg/mL) on peptide
productions. The experimental results were summarized in Table
1. The results could be elucidated by the following non-linear
regression model using the SPSS software (IBM SPSS version 21).
Y=111.537-0.699 × X1-0.908 × X2-1.614 × X3+0.047 × X1 × X1+0.190
× X2 × X2+0.003 × X3 × X3-0.252 × X1 × X2+0.045 × X1 × X3+0.042 × X2 × X3
In this equation, Y refers to peptide production yield (%), X1 to
the proteinase-K concentration (mg/mL), X2 to the reaction
incubation time (h), and X3 to the reaction temperature (°C). This
yield model was highly significant (p <0.01) with an important
determination value of (R2= 0.917) confirming that 91.7% of the
inconsistency in the response could be explained by the above
model. This equation was then investigated by means of “F”
statistical analysis by ANOVA to test the goodness of fit. The “F”
value (111.396), with a very low probability value, (p <0.001)
demonstrated that this equation was highly significant. Indeed,
the higher of the “F” value is the more significant deduced model
becomes. Then, the adjusted coefficient of determination (Ra2=
0.913) proved a better consistency and precision of the approved
assays [2,30].
The studies of the following equation also displayed that
peptide production depended ultimately on the linear and
quadratic terms of the three selected parameters. In this
model, the interactions between temperature with, respectively,
proteinase-K concentration (X1, X3) and reaction incubation
time (X2, X3) show a positive effect on hydrolysis yield, whereas
proteinase K concentration and reaction time of incubation
(X1, X2) shows a negative effect. The interactions between the
parameters could be well visualized by plotting the 3D response
surface designs. Consequently, the response plots were done
by varying the two variables while the third one was kept at its
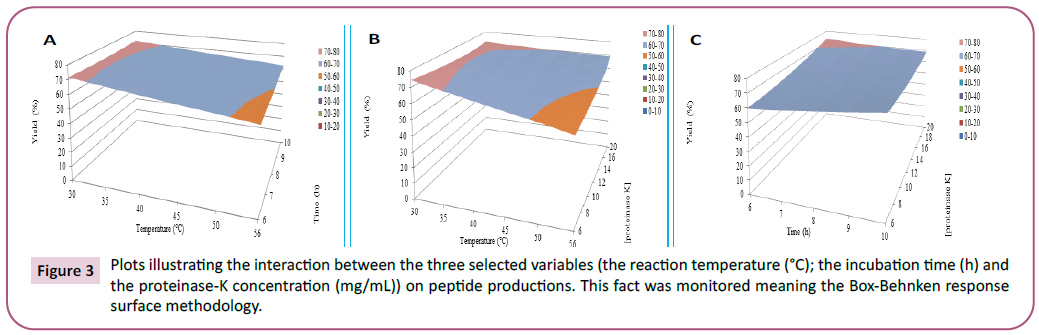
middle level as indicated in Figure 3.
Figure 3: Plots illustrating the interaction between the three selected variables (the reaction temperature (°C); the incubation time (h) and the proteinase-K concentration (mg/mL)) on peptide productions. This fact was monitored meaning the Box-Behnken response surface methodology.
These surface response plots exhibited a significant curvature in
curves suggesting that these variables were interdependent at
different levels. The peptide production level increased when the proteinase-K content increased but it decreased at a high level of
temperature. Additionally, the optimal conditions were found in
the high level of the reaction incubation time values. The validity
of the polynomial equation was verified by doing the optimum
conditions: 20 mg/mL of proteinase-K concentration, a 10 h of
reaction time incubation, and 30°C of incubation temperature.
The predicted peptide production was 79.83%. This production
was validated through three experiments at an optimal condition
whose results were 81%, 82%, and 83%. The high correlation between the predicted and the experimental values displayed
the validity of the response fitted model. The peptide yield was
approximately 2.66 times higher than the one obtained before
optimization. Certainly, controlling the physical parameters of the
hydrolyzing dramatically enhanced the peptide production.
Chromatographic studies and characterizations
of the hydrolysate
After hydrolyzing the prepared Rubisco solution meaning proteinase-K, we proceed to the separation of the potential
released peptides. In fact, the alfalfa Rubisco was loaded to
reversed-phase high performance liquid chromatography analysis
in order to evaluate its purity and the potential effects of the
extraction process on protein quality. To do this, the hydrolysate
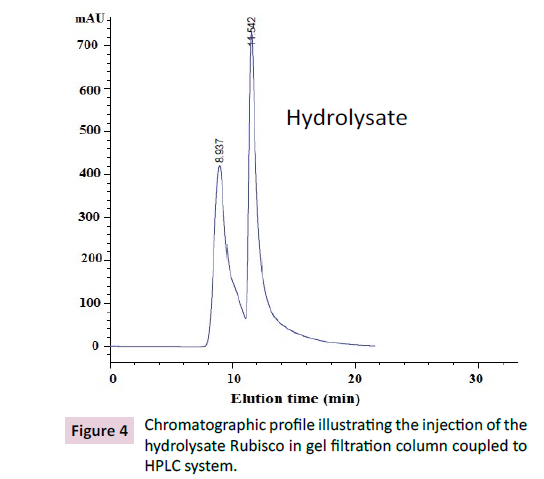
was injected to gel filtration column coupled to HPLC system. The
result is followed in Figure 4 illustrating the chromatographic
profile.
Figure 4: Chromatographic profile illustrating the injection of the
hydrolysate Rubisco in gel filtration column coupled to
HPLC system.
The presence of two peaks at an elution time of 9 min and 11.5
min was taken as an indicator for the presence of Rubisco in the
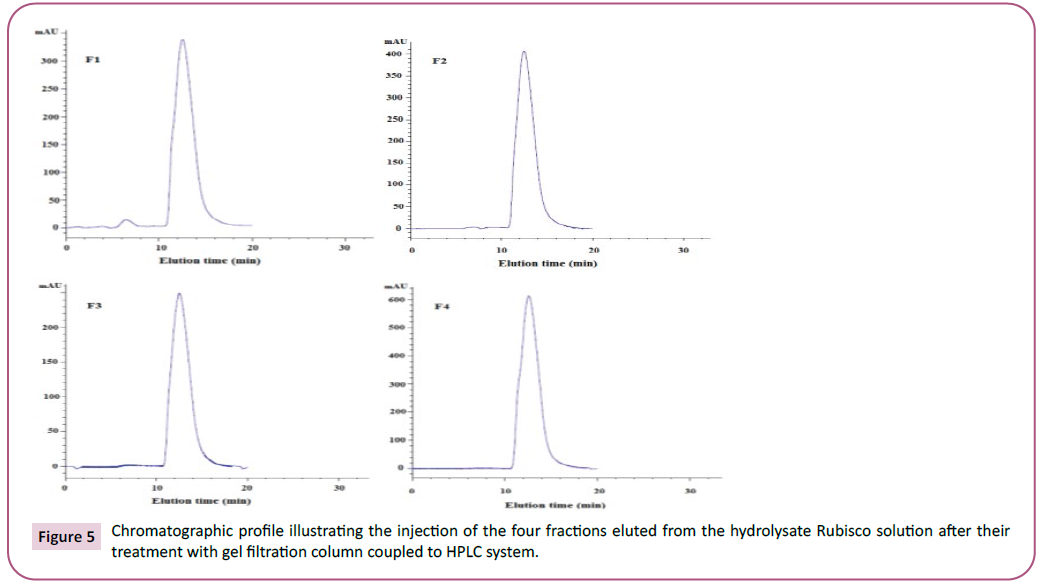
sample [31]. Indeed, four fractions were recuperated (named F1,
F2, F3 and F4, respectively) and re-injected on the same column–
HPLC system, as shown in Figure 5. The described eluted fractions
will be used as plausible tools for some of biotechnological
applications related to para-pharmaceutical and agrofood fields.
Figure 5: Chromatographic profile illustrating the injection of the four fractions eluted from the hydrolysate Rubisco solution after their treatment with gel filtration column coupled to HPLC system.
Radical-scavenging system assessment
For more characterizations, the separated fractions were used
to measure some physiological parameters such as CAT, SOD and
DPPH. Indeed, those parameters were monitored in the case of
the crude extract and also in the case of the peptide fractions.
Indeed, Table 3 follows that the CAT activity level seems to be the
same on the crude extract and the F4 fraction. This finding pleads
for the fact that this enzymatic activity is encoded, for probably,
by F4. In another hand, the SOD activity monitored on the crude
extract is not arised in any one of those described fractions (F1,
F2, F3 and F4). In addition, the DPPH appears in all the eluted
fractions and we can’t attribute this physiological parameter to
one of them. Eventually, the described eluted fractions are able
to be used as effectors in various fields.
| |
CAT (U/mg protein) |
SOD (U/mg protein) |
DPPH (%) |
| Crude Extract |
79,73 |
1289,13 |
35,34 |
| F1 |
18,4 |
30,54 |
78,87 |
| F2 |
2,3 |
26,15 |
56,89 |
| F3 |
11,5 |
43,65 |
49,13 |
| F4 |
57,5 |
19.35 |
37,93 |
Table 3: Illustration of the assessment of the CAT, SOD and DPPH levels of the crude extract and the described eluted fractions (F1, F2, F3 and F4).
According to Nelson SK, et al. [32], a study was done on healthy
human subjects whom were injected by a mixture of extracts
of five medicinal plants after induction of SOD and CAT. This
experiment was based on the results made on rodents with
evidence of decreasing lipid peroxidation. Findings showed that
modest induction of the catalytic antioxidants SOD and CAT
may be a much more effective approach than supplementation
with antioxidants (such as vitamins C and E) that can, at best,
stoichiometrically scavenge a very small fraction of total oxidant
production. In addition to that, others findings demonstrate that
the altered energy balance in High fat diet fed mitochondrial
catalase mice protected them diacylglycerol accumulation,
protein kinase C activation and impaired muscle insulin signaling
[33]. According to those data, we will be look for the plausible
use of the catalase capability of our biological matrix as a
pharmaceutical or/and para-pharmaceutical product.
Conclusion
Based on those findings, we conclude that the Alfalfa Ribulose-
1,5-bisphosphate carboxylase/oxygenase will be an excellent
candidate protein in the future so as to contribute to the
improvement of various biotechnological processes related to
the agro-food industry as additive and also for their capability as
clinical and also as pharmacological effector.
Acknowledgment
This work was in part financially supported by a grant from the
Tunisian Ministry of Higher Education and Scientific Research
contract program_2015-2018 and also the program_2013-2022
CBS-LBAP/code: LR15CBS03 and Institute of Arid Lands-
Medenine: Arid and Oases Cropping Laboratory (IRA-LACO).
References
- Raes M, Planque M, Marbaix H, Gillard N, Lecrenier MC, et al. (2018) Mass Spectrometry based Approaches for Analysing Proteins in Processed Food and Feed. Clinics in Oncology 3: 2-18.
- Nehal F, Sahnoun M, Dab A, Sebaihia M, Bejar S, et al. (2019) Production optimization, characterization, and covalent immobilization of a thermophilic Serratia rubidaea lipase isolated from an Algerian oil waste. Molecular biology reports 46: 3167-3181.
- Billen G, Lassaletta L, Garnier J, Le Noë J, Aguilera E, et al. (2019) Opening to distant markets or local reconnection of agro-food systems? Environmental consequences at regional and global scales. Agroecosystem Diversity 391-413.
- Steinfeld H, Gerber P, Wassenaar T, Castel V, Rosales M, et al. (2006) Livestock's long shadow: environmental issues and options. Food & Agriculture Org.
- Raschka A, Carus M (2012) Industrial material use of biomass basic data for Germany. Europe and the world, nova-Institute.
- Aiking H (2011) Future protein supply. Trends in Food Science & Technology 22: 112-120.
- Day L (2013) Proteins from land plants–potential resources for human nutrition and food security. Trends in Food Science & Technology 32: 25-42.
- Davys M, Richardier F, Kennedy D, de Mathan O, Collin S, et al. (2011) 18 Leaf Concentrate and Other Benefits of Leaf Fractionation. Combating Micronutrient Deficiencies: Food-based Approaches p. 338.
- Agarwal A, Mathur B, Joshi P (2013) Efficacy of leaf concentrate as micronutrient fortifier in the supplementary nutrition of Integrated Child Development Services (ICDS). International Journal of Nutrition and Metabolism 5: 98-105.
- Kobbi S, Bougatef A, Balti R, Mickael C, Fertin B, et al. Purification and recovery of RuBisCO protein from alfalfa green juice: antioxidative properties of generated protein hydrolysate. Waste and Biomass Valorization 8: 493-504.
- Julier B, Semiani Y, Laouar M (2010) Genetic Diversity in a Collection of Lucerne Populations from the Mediterranean Basin Evaluated by SSR Markers, Sustainable use of genetic diversity in forage and turf breeding. Springer 107-112.
- Rafińska K, Pomastowski P, Wrona O, Górecki R, Buszewski B (2017) Medicago sativa as a source of secondary metabolites for agriculture and pharmaceutical industry. Phytochemistry Letters 20: 520-539.
- Raven JA (2013) Rubisco: still the most abundant protein of Earth?. New Phytologist 198: 1-3.
- Gaweł E, Grzelak M, Janyszek M (2017) Lucerne (Medicago sativa L.) in the human diet—Case reports and short reports. Journal of Herbal Medicine 10: 8-16.
- Chen W, Qiu Y (2003) Leaf protein’s utilization status and its prospect. Food Science 24: 158-161.
- Udenigwe CC, Gong M, Wu S (2013) In silico analysis of the large and small subunits of cereal RuBisCO as precursors of cryptic bioactive peptides. Process Biochemistry 48: 1794-1799.
- Kohler G, Knuckles B (1977) Edible protein from leaves. Food Technology.
- Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, et al. (2007) Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 71: 576-599.
- Telek L, Graham HD (1983) Leaf protein concentrates.
- Hamm HE (2001) How activated receptors couple to G proteins. Proceedings of the National Academy of Sciences 98: 4819-4821.
- Incamps A, Hély‐Joly F, Chagvardieff P, Rambourg JC, Dedieu A, et al. (2005) Industrial process proteomics: alfalfa protein patterns during wet fractionation processing. Biotechnology and Bioengineering 91: 447-459.
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72: 248-254.
- Laemmli UK, Favre M (1973) Maturation of the head of bacteriophage T4: I. DNA packaging events. Journal of Molecular Biology 80: 575-599.
- Hiidenhovi J, Hietanen A, Mäkinen J, Huopalahti R, Ryhänen EL (2005) Hydrolysis of ovomucin by different enzymes, Proceedings of the XVII European Symposium on the Quality of Poultry Meat and XI European Symposium on the Quality of Eggs and Egg Products, Golden Tulip Parkhotel Doorwerth, Doorwerth, Netherlands, 23-26 May 2005, World's Poultry Science Association (WPSA) 251-256.
- Bersuder P, Hole M, Smith G (1998) Antioxidants from a heated histidine-glucose model system. I: Investigation of the antioxidant role of histidine and isolation of antioxidants by high-performance liquid chromatography. Journal of the American Oil Chemists' Society 75: 181-187.
- Hsouna AB, Saad RB, Zouari N, Romdhane WB, Brini F, et al. (2019) Stress associated protein from Lobularia maritima: Heterologous expression, antioxidant and antimicrobial activities with its preservative effect against Listeria monocytogenes inoculated in beef meat. International Journal of biological Macromolecules.
- Ellis RJ (1979) The most abundant protein in the world. Trends in biochemical sciences 4: 241-244.
- Müller KD, Salnikow J, Vater J (1983) Amino acid sequence of the small subunit of D-ribulosebisphosphate carboxylase/oxygenase from Nicotiana tabacum. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology 742: 78-83.
- Gibbs BF, Zougman A, Masse R, Mulligan C (2004) Production and characterization of bioactive peptides from soy hydrolysate and soy-fermented food. Food Research International 37: 123-131.
- Khuri A, Littell R (1987) Exact tests for the main effects variance components in an unbalanced random two-way model.Biometrics 545-560.
- Trovaslet M, Kapel R, Ravallec‐Plé R, Mouni F, Clarisse M, et al. (2007) Secretagogue and bacteriostatic active fractions derived from a peptic hydro‐lysate of alfalfa RuBisCO small purified subunit. Journal of the Science of Food and Agriculture 87: 534-540.
- Nelson SK, Bose SK, Grunwald GK, Myhill P, McCord JM (2006) The induction of human superoxide dismutase and catalase in vivo: A fundamentally new approach to antioxidant therapy. Free Radical Biology and Medicine 40: 341-347.
- Lee HY, Lee JS, Alves T, Ladiges W, Rabinovitch PS, et al. (2017) Mitochondrial-Targeted Catalase Protects Against High-Fat Diet–Induced Muscle Insulin Resistance by Decreasing Intramuscular Lipid Accumulation. Diabetes 66: 2072-2081.