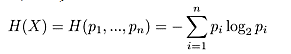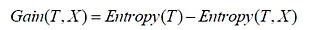Research Article - (2022) Volume 6, Issue 12
An Algorithm for Notifiable Disease Modeling and Prediction using Artificial Intelligence Techniques: A Case of Kenya
Nicodemus Nzoka Maingi1*,
Ismail Ateya Lukandu1 and
Matilu Mwau2
1Department of Computing and Engineering Sciences, Strathmore University, Nairobi, Kenya
2Department of Computing and Engineering Sciences, Kenya Medical Research Institute, Nairobi, Kenya
*Correspondence:
Nicodemus Nzoka Maingi, Department of Computing and Engineering Sciences, Strathmore University, Nairobi,
Kenya,
Email:
Received: 25-Jul-2022, Manuscript No. IPJCGH-22-14022;
Editor assigned: 27-Jul-2022, Pre QC No. IPJCGH-22-14022(PQ);
Reviewed: 10-Aug-2022, QC No. IPJCGH-22-14022;
Revised: 08-Dec-2022, Manuscript No. IPJCGH-22-14022(R);
Published:
16-Dec-2022, DOI: 10.36648/2575-7733.6.12.57
Abstract
Background: The disease outbreak management operations of most countries (notably Kenya)
present numerous novel ideas of how to best make use of notifiable disease data to effect proactive
interventions. Notifiable disease data is reported, aggregated and variously consumed. Over the
years, there has been a deluge of notifiable disease data and the challenge for notifiable disease data
management entities has been how to objectively and dynamically aggregate such data in a manner
such as to enable the efficient consumption to inform relevant mitigation measures. Various models
have been explored, tried and tested with varying results; some purely mathematical and statistical,
others quasi mathematical cum software model driven.
Methods: One of the tools that have been explored is Artificial Intelligence (AI). AI is a technique that
enables computers to intelligently perform and mimic actions and tasks usually reserved for human
experts. AI presents a great opportunity for redefining how the data is more meaningfully processed
and packaged. This research explores AI’s Machine Learning (ML) theory as a differentiator in the
crunching of notifiable disease data and adding perspective. An algorithm has been designed to test
different notifiable disease outbreak data cases, a shift to managing disease outbreaks via the
symptoms they generally manifest. Each notifiable disease is broken down into a set of symptoms,
dubbed symptom burden variables, and consequently categorized into eight clusters: Bodily, gastro
intestinal, muscular, nasal, pain, respiratory, skin, and finally, other symptom clusters. ML’s decision
tree theory has been utilized in the determination of the entropies and information gains of each
symptom cluster based on select test data sets.
Results: Once the entropies and information gains have been determined, the information gain
variables are then ranked in descending order; from the variables with the highest information gains
to those with the lowest, thereby giving a clear cut criteria of how the variables are ordered. The
ranked variables are then utilized in the construction of a binary decision tree, which graphically and
structurally represents the variables. Should any variables have a tie in the information gain rankings,
such are given equal importance in the construction of the binary decision tree. From the presented data, the computed information gains are ordered as; gastro intestinal, bodily, pain, skin, respiratory,
others. Muscular and finally nasal symptoms respectively. The corresponding binary decision tree is
then constructed.
Conclusions: The algorithm successfully singles out the disease burden variable(s) that are most
critical as the point of diagnostic focus to enable the relevant authorities take the necessary, informed
interventions. This algorithm provides a good basis for a country’s localized diagnostic activities driven
by data from the reported notifiable disease cases. The algorithm presents a dynamic mechanism that
can be used to analyze and aggregate any notifiable disease data set, meaning that the algorithm is
not fixated or locked on any particular data set.
Keywords
Artificial intelligence; Decision tree; Disease burden; Disease surveillance; Disease
symptom; Decision tree theory; Entropy; Information gain
Abbreviations
AI: Artificial Intelligence; Epi-Week: Epidemiological Week; CDC: Centre for Disease
Control and Prevention; DDSR: Division of Disease Surveillance and Response; ML: Machine Learning;
USAID United Stated Agency for International Development; WHO: World Health Organization.
Introduction
Gastric disease surveillance is an information based activity
involving the collection, analysis and interpretation of large
volumes of disease outbreak data from a variety of sources in
order to inform and drive objective and informed
intervention. The Disease Surveillance and Response Unit
(DSRU) is the entity mandated (in Kenya) to monitor and
undertake response and mitigation measures in the event of a
notifiable disease outbreak; a notifiable disease refers to any
disease in a country or community whose occurrence must be
reported to the authorities. Each time a notifiable disease is
reported, the DSRU undertakes the necessary response
activities. In Kenya, disease outbreaks are mostly tackled from
two perspectives; reactive measures in the event a notifiable
disease outbreak is reported, mitigating steps are only
undertaken in response to the particular incident(s) to
minimize the potential consequent adverse effects; not much
is learned or information utilized in the aftermath that could
meaningfully, incrementally and objectively inform future
outbreaks and; proactive measures anticipatory measures are
put into play such that should an outbreak occur or recur, its
adverse effects are greatly minimized with health personnel
taking informed, premeditated and experience-driven steps as
a better approach to empower the health personnel be better
prepared to cope with every subsequent outbreak. The
infectious diseases of the past have been known to have
included some of the most contagious and feared plagues of
the past, with new strains continuing to emerge over time;
this warrants a widely and greatly co-operative and proactive
approach even when the disease outbreak responses and
intervention efforts remain the prerogative of the concerned
national government. Global partners (such as the Centre for
Disease Control and prevention (CDC), the United Stated
Agency for International Development (USAID), the World
Health Organization (WHO) among others) have also been
seen to play a great role by working in close collaboration to
offer the much needed medico-technical and social support
from its battery of experienced and seasoned teams cutting across numerous medical specialities and vast geopolitical
backgrounds. To enable each country’s concerned teams
better manage its disease outbreaks more efficiently, a
notifiable disease list and its epidemiological week (epi week)
must be defined; an epi week is a weekly period in a country
within which notifiable disease outbreak data must be
recorded and reported to the relevant health authorities.
Kenya’s epi week runs from monday through sunday. The
efforts to manage disease outbreaks have become a very
complex endeavor; historically, it was easier due to smaller
populations and the limited, minimized yet localized cross
border and cross territorial movements and interactions that
curtailed the cross pollination or dissemination of infectious
diseases the concerned population may have been harboring
this has greatly changed in the advent of globalization. The
effects of globalization have brought forth new dynamic risk
factors in disease spread and management. Such factors
include: Faster and easy cross-border movements of people
and animals, making diseases spread faster for instance,
urbanization remains one of the greatest factors of disease
spread: New urban settlements and availability of a huge
community of commuting skilled and readily available labour
across geopolitical boundaries having the ability to create
some infection epicenters that if not well managed, could
easily become incubators for new epidemics, and zoonotic
diseases, which can spread in a more rapid manner, quickly
elevating them to global levels of interest and concern. Next
comes means of transporting goods or parcels. The efficient
and rapid movement of goods also presents a possibility of
enabling and enhancing the spread of diseases since the
goods may be harboring and transporting whatever existing
disease strain to wherever they are transported or delivered.
Additionally, there is also the new, modern practice of
families frequently eating out where they get more exposed
to different infectious disease strains, among other
exposures. Suddenly, one nation’s (seemingly localized)
epidemic challenges quickly become other nations, regions
and partners health concerns pathogens are not known to
commonly follow or respect geopolitical and human
boundaries. Additionally, in economic and industrial competitive terms, other factors could also kick in for
instance, the economic empowerment or disempowerment of
the notifiable disease affected populations when skilled,
experienced and knowledgeable working personnel get
grossly affected by a disease. The push and pull factors for
disease surveillance also touch on the socio economic
activities of a nation; disease outbreaks have been known to
decimate the knowledgeable, skilled and able bodied working
populations of any nation to a point of economic near
standstill if not total collapse. Further, it is has been observed
that the progression or retrogression of the economic
wellbeing of a community can now be greatly tied to proper
disease outbreak management; if the adverse effects slow
down economic activity, then all measures, (including the
improvement of the health infrastructure and the response
and mitigation apparatus of a country) must be called upon to
prevent or deal with the adversity of the disease outbreaks.
To combat such disease strains, concerted efforts and clear
cut strategies need to be employed; the enhanced use of ICT
software and tools has been seen as a great driver and
catalyst to enable the quick aggregation, packaging and
dissemination of disease data through to the relevant
personnel for easier, faster and better informed interventions.
The disease outbreak data used here is subjected to AI’s
machine learning theory. Machine learning is a technique that
provides systems with the ability to automatically learn and
improve from experience. Whilst traditional disease outbreak
management assumes the method of relying on past disease
data that is seen to point towards what infectious disease
strains manifest, this research looks to dig deeper. Using AI,
the researcher hopes to drive a different perspective to notifiable disease outbreak management of the two disease
outbreak management perspectives outlined earlier, the
researcher looks to build on the proactive disease outbreak
measures. The main driving question or hypothesis here is
whether a different approach could be employed to the
processing and packaging of notifiable disease data in order
to better inform and drive proactivity in the disease
surveillance and response practice [1-8].
Materials and Methods
The methodology used here employs various
techniques; quantitative and qualitative research analysis
blended with evolutionary and iterative prototyping. The
C4.5 decision tree theory in artificial intelligence has been
used in the diagnostic analysis efforts, with the computed
information gains consequently becoming reliable
determinants in informing the structure of the resultant
binary decision tree(s). Post validation, the algorithm could
be further applied to the general notifiable disease list across
many counties and regions to handle the variation of the
disease outbreak footprints as an additional test measure of
the algorithm’s efficacy; it is expected that any challenges
experienced in the process of the development of the
algorithm will be used as a basis for future improvement
and to inform policy development and assist in better
planning efforts [9-11].
The eight symptom clusters adopted are listed below Table 1.
| Symptom cluster code |
Symptom cluster |
Brief description |
| B |
Bodily |
Those symptoms that are generally manifested through the general human body organs and parts e.g. fever. |
| G |
Gastro-Intestinal |
Those symptoms that are generally manifested through the human body’s digestive system e.g. vomiting, running stomach etc. |
| M |
Muscular |
Those symptoms that are manifested via the human body’s muscular tissues. |
| N |
Nasal |
Those symptoms that are generally manifested through the human nasal cavity e.g. running nose, sneezing etc. |
| P |
Pain |
Those symptoms that manifest in form of various forms of human body pain e.g. headache. |
| R |
Respiratory |
Those symptoms that manifest through the human body’s respiratory processes or apparatus e.g. shortness of breath, coughing. |
| S |
Skin |
Those symptoms that manifest through a human body’s skin tissue e.g. skin rush, skin peeling or inflammation. |
| O |
Other |
Those symptoms that generally fall outside the other seven defined symptom clusters e.g. blurred eye sight. |
Table 1: Disease symptom clusters legend.
Entropy: Equation 1-entropy computation.
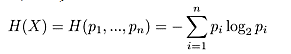
Information gains: Equation 2 information gains computation.
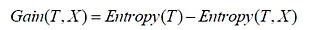
Once the information gains are computed, their rankings are
used to determine the order of the symptom variable clusters
in the construction of the binary decision tree, yielding a
structure that helps to graphically and visually break down the
notifiable disease outbreak data into a meaningful form to
guide intervention and proactive action. The ranked symptom
burden variables can assist the health personnel in easily
mapping what disease symptom variable (s) to lay emphasis
upon in their efforts to combat outbreaks. This means that
there is a deviation from the traditional practice of the focus
being laid upon the singular diseases themselves; the
combating of disease outbreaks would mainly be driven by
the symptom cluster variables i.e. it is possible to focus on
only those diseases that manifest certain symptom cluster
variables that are highly ranked via the algorithm using the
computed information gains. Thus, the planning and
mitigating measures would mainly be on the diseases
symptom variables, and not necessarily the raw disease(s)
themselves. Each notifiable disease data set follows the
information gains ranking. For instance, if the Pain symptom
variable ranks first in the information gains computation, then
it will become the root node in the resultant binary decision
tree. The rest of the symptom variables will follow
accordingly. If two or more variables tie in the information
gain ranks, then they shall jointly be part of a leaf node (or the
root node, if they tie on rank one) as the decision tree gets
defined and constructed. For purposes of the validation of the
algorithm, the researcher chose to use the C4.5 machine
learning binary decision tree theory in the computation of the
entropies and information gain values prior to the definition
and construction of the binary decisions trees. The data used
in the validation of this algorithm is aggregated from Kenya’s
Nairobi County over the 2015–2018 notifiable disease
reporting period.
Below is the algorithmic process flow of the various activities
(Figure 1).
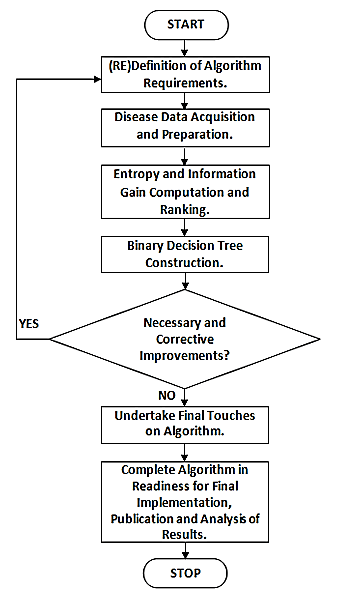
Figure 1: Algorithmic process flow.
Results
The information gain scores tabulated here are derived from
the data sets prepared from the primary data.
The information gain scores are ranked from the largest to the
smallest; with the highest information gain score pointing to
a particular symptom variable(s) that is the most critical in the
decision tree construction, whilst the smallest information
gain score points to the variable that is the least important
as a binary decision tree determinant variable (Table
2) [12-15].
| Nairobi county information gains disease symptom burdens |
| C4.5 Technique data |
| Gain (decision, variable) and gain rankings |
| Overall aggregated data |
| Variable |
B |
G |
M |
N |
O |
P |
R |
S |
| Information gain |
4.3496 |
4.4366 |
1.0144 |
0.7654 |
2.206 |
3.4801 |
2.3451 |
2.8691 |
| Rankings |
2 |
1 |
7 |
8 |
6 |
3 |
5 |
4 |
Table 2: Nairobi county 2015–2018 aggregated information gain scores and rankings.
The binary decision tree shall then be constructed according
to the information gain rankings tabulated above (Figures 2
and 3).
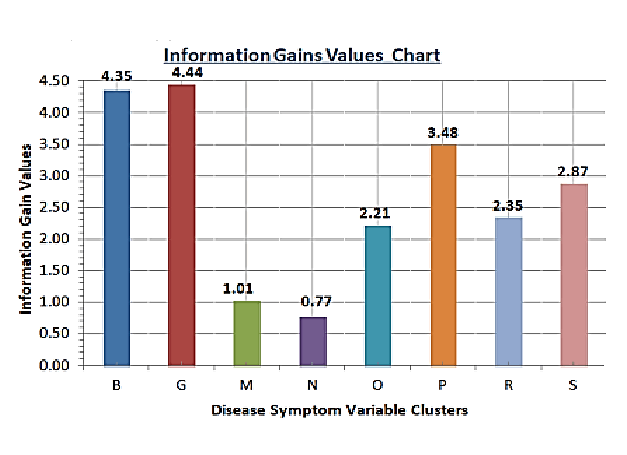
Figure 2: Symptom variable clusters information gains chart.
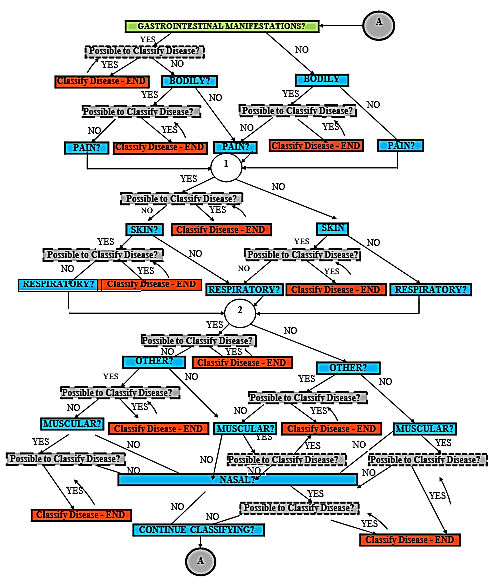
Figure 3: C4.5 Nairobi county binary decision tree.
Discussion
It is a prerogative of every nation to focus on the
strengthening of its public health infrastructure to protect its
citizen’s health, and especially in combating disease
outbreaks. Thus, all disease outbreak factors can easily be
dealt with. This research looks to present a good push in
innovating new approaches and methodologies in the
development of a proactive, early warning system in the
response and intervention efforts to support some medium
and long term mechanisms for the processing of the disease
data with the focus on specific trends to inform policy
development and planning, thereby boosting decision making
at the DSRU in collaboration with other concerned partners.
The algorithm demonstrates interesting results of the eight
disease symptom burden cluster variables, the gastro
intestinal variable emerges as the most prominent, having
registered an information gain score of 4.4366. It goes on to
form the root node (the first node of a binary decision tree). It
is closely followed by the bodily symptom variable with an
information gain score of 4.3496. The others follow in the
following order (based on the information gain scores): Pain
(3.4801), skin (2.8691), respiratory (2.3451), others (2.2060),
muscular (1.0144) and finally nasal manifestations (0.7654).
With the gastro intestinal variable emerging as the most
highly ranked variable, this means that the disease mitigation
efforts and focus should be laid upon those diseases that
manifest any gastro intestinal symptoms. Once these have
been exhaustively addressed, next will come those with
bodily symptoms. Consideration should be taken right from
the most highly ranked symptom variable cluster to the
lowest in order to objectively guide the diagnostic
preparedness of an entity (be it a nation, a province, county
or any other geographical demarcation possible) the
proposed shift here means there is a deviation from the
traditional diagnostic practice of focus and diagnostic
emphasis being laid upon diseases individually; instead, each
disease is defined as a set of symptoms within the defined
disease symptom clusters. The algorithm can then be applied;
simply determine the information gains of each of the
variable, the rankings and consequent variable classification
and finally, the binary decision tree construction. Learning is a
multi-faceted occurrence, with learning processes involving
the acquisition of new declarative knowledge, the
development of motor and sensitive skills through instruction
and practice ordering of the new knowledge. Tools to drive such new knowledge include artificial intelligence branches
such as machine learning, as utilized in this research. The
essence of this algorithm is to derive a seemingly localized
diagnostic framework to enable local medical personnel easily
manage disease outbreaks by predicting what disease
symptom variables should be given priority in the fight against
outbreaks. This approach assumes that in order to manage
disease outbreaks on an ongoing basis, all the diseases’
should be classifiable within the eight symptom clusters. The
algorithm then goes on to cluster the diseases based on their
most critical symptom burdens. Emphasis is laid on the
disease symptom variables i.e. the disease(s) that manifest(s)
a certain highly ranked symptom variable is given more
prominence in the diagnostics and interventional process. The
algorithm’s ranking of symptom variables is purely data driven i.e. as new data is posted, the symptom variables’ information
gains are expected to keep changing and assuming new ranks,
thereby dynamically changing the order of importance of the
symptom variables of focus. As such, the fight against disease
outbreaks focuses not on the diseases, but by the symptoms
that drive these diseases of great importance is the
management of disease outbreaks by providing an objective
basis for crunching and aggregating the data in a novel and
objective way to easily inform decision making.
Conclusion
In conclusion, it has been demonstrated that the disease
management efforts of an entity can be purely driven by the
disease data presented aided by the just defined and
validated algorithm. This research study ends up creating a
case for disease diagnostics mainly using symptom burden
variables. Notably, a case for the machine learning driven
algorithm has been presented together with its validation
process. Additionally, the algorithm has been used in the
computation of the information gains and their rankings.
Finally, the just defined, computed and ranked information
gains have been shown to form a basis for the definition and
construction binary decision tree. In the end, the algorithm
has been designed, constructed and validated. The whole
process easily enables the disease outbreak management
exercise of any local authority be home grown i.e. The basis of
the disease outbreak management can be guided and driven
by the local disease data being captured and continuously
crunched to keep the disease diagnostics exercise as fluid and
as objective as the data that drives it.
Competing Interests
The researcher wishes to declare that there are no known
competing interests to this research article.
Acknowledgements
The researcher wishes to acknowledge the support received
from the Kenya Medical Research Institute (KEMRI) as well as
that from Strathmore University.
References
- Baker EL, Koplan JP (2002) Strengthening the Nation’s Public Health Infrastructure: Historic Challenge, Unprecedented Opportunity. Health Aff (Millwood). 21(6):15-27.
[Crossref][Googlescholar][Indexed]
- Bernardo TM, Rajic A, Young I, Robiadek K, Pham MT, et al. (2013) Scoping Review on Search Queries and Social Media for Disease Surveillance: A Chronology of Innovation. J Med Internet Res. 15(7):147.
[Crossref][Googlescholar][Indexed]
- Brownstein JS, Freifeld CC, Madoff LC (2009) Digital Disease Detection Harnessing the Web for Public Health Surveillance. N Engl J Med. 360(21):2153-2157.
[Crossref][Googlescholar][Indexed]
- Centre for Disease Control (CDC) (2012). Summary of Notifiable Diseases (SND)-United States, 2010. Morbidity and Mortality Weekly Report (MMWR). 59(53):1
[Googlescholar][Indexed]
- Childs JE, Krebs JW, Real LA, Gordon ER (2007) Animal based National Surveillance for Zoonotic Disease: Quality, Limitations, and Implications of a Model System For Monitoring Rabies. Prev Vet Med. 78: 246-261.
[Crossref][Googlescholar][Indexed]
- Kulldorff M (2001) Prospective Time Periodic Geographical Disease Surveillance Using a Scan Statistic. J R Stat Soc Ser A Stat Soc. 164(1):61-72.
[Crossref][Googlescholar][Indexed]
- Mack A, Choffnes ER, Relman DA. Infectious Disease Movement in a Borderless World: Workshop Summary. Institute of Medicine, Board on Global Health. National Academies Press. Washington Dc, USA. (2010). 1-322.
[Googlescholar]
- Martinez L (2000) Global Infectious Disease Surveillance. Int J Infect Dis. 4: 222-228.
[Crossref][Googlescholar]
- Moncayo A, Silveira AC (2009) Current Epidemiological Trends for Chagas Disease in Latin America and Future Challenges In Epidemiology, Surveillance and Health Policy. Memórias do Instituto Oswaldo Cruz. 104: 17-30.
[Crossref][Googlescholar][Indexed]
- Neiderud CJ (2015) How Urbanization Affects the Epidemiology of Emerging Infectious Diseases. Infect Ecol Epidemiol. 5(1):27060.
[Crossref][Googlescholar][Indexed]
- Nsubuga P, Nwanyanwu O, Nkengasong JN, Mukanga D, Trostle M (2010) Strengthening Public Health Surveillance and Response Using the Health Systems Strengthening Agenda in Developing Countries. BMC Public Health. 10(1):1-5.
[Googlescholar]
- Wagner MM, Tsui FC, Espino JU, Dato VM, Sittig DF, et al. (2001) The Emerging Science of Very Early Detection of Disease Outbreaks. J Public Health Manag Pract. 7(6):51-59.
[Crossref][Googlescholar][Indexed]
- Weinberg M, Waterman S, Lucas CA, Falcon VC, Morales PK, et al. (2003) The US-Mexico Border Infectious Disease Surveillance Project: Establishing Bi-National Border Surveillance. Emerg Infect Dis. 9(1):92-97.
[Crossref][Googlescholar][Indexed]
- World Health Organization (WHO) (2006) Communicable Disease Surveillance and Response Systems: Guide to Monitoring and Evaluating. The U.S. Government and the World Health Organization, USA. 1-90.
[Googlescholar]
- Zhong Y, Oh S, Moon HC (2021) What Can Drive Consumers Dining Out Behavior in China and Korea during the COVID-19 Pandemic? Sustainability. 13(4): 1724.
[Googlescholar][Indexed]
Citation: Maingi NN, Lukandu IA, Mwau M (2022) An Algorithm for Notifiable Disease Modeling and Prediction using
Artificial Intelligence Techniques: A Case of Kenya. J Clin Gastroenterol Hepatol. 6:57.
Copyright: © 2022 Maingi NN, et al. This is an open-access article distributed under the terms of the Creative Commons
Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author
and source are credited.