Keywords
Gene expression; Transcription; ARF-19; Arabidopsis
Introduction
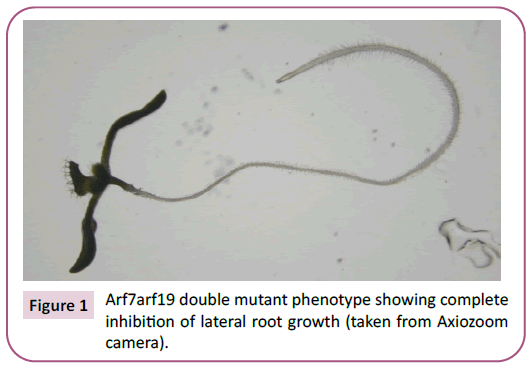
The plant hormone auxin plays a central role in plant growth and development through the control of the expression of many genes. Auxin response factors (ARFs) are transcriptional factors that bind to DNA and activate the expression of genes in response to auxin signaling. ARFs are required to form homodimers in order to bind to promoter DNA and activate transcription of a gene [1]. The ARF dimers specifically bind to sequences known as auxin response elements (AUxREs) in the promoter region of the genes. Without the presence of auxin, ARFs form heterodimeric proteins with Auxin/Indole-3-Acetic Acid (Aux/IAA) repressor proteins, which then bind to the promoters and prevent gene expression from occurring. Auxin is crucial for gene expression because it promotes ARFs to form the homodimers by facilitating the degradation of the repressor proteins by ubiquitin ligase [2]. This study was specifically focused on ARF-19 in an attempt to uncover a downstream target of this transcription factor and further understand how plant transcription factors work (Figure 1).
Figure 1: Arf7arf19 double mutant phenotype showing complete inhibition of lateral root growth (taken from Axiozoom camera).
Using genomic data of the plant Arabidopsis thaliana, we identified a potential target whose expression was observed in budding yeast, Saccharomyces cerevisiae.In A. thaliana, ARF-7 and ARF-19 are known to promote lateral root formation in an auxin-mediated response pathway. As auxin acts on many transcription factors in this plant (Arabidopsis has 23 ARFs), it makes it harder to determine the expression/function of one gene. As seen in previous research, either ARF-7 or ARF-19 mutants are not sufficient to completely inhibit lateral root growth; only the double mutant where both transcription factors were not expressed showed a severely inhibited growth phenotype [3]. Yeast is an ideal organism for this study as it does not possess any endogenous auxin, which allowed us to isolate the expression of one gene activated by ARF-19 (Figure 2). The Arabidposis gene that we ultimately analyzed as a potential downstream target was AT1G51950, which is more commonly referred to as IAA18. At first, we selected the AT5G64110 gene due its role in oxidative stress defense, but the inability to construct a plasmid with its promoter forced us to change targets to IAA18. Here, we describe the process of analyzing our potential downstream target of ARF-19, and our results can help provide a better understanding of how plant transcription factors work and what genes ARF-19 acts on to promote Arabidopsis growth and development.
Figure 2: Arf19 mutant phenotype taken using the Axiozoom camera. The single mutant still shows lateral root growth, indicating there are many Arfs in plants that stimulate growth.
Methods
Gene identification and selection
RNA-seq. and microarray data were provided for us from Lewis et al. to identify key transcription factors and to get the promoter sequences of potential ARF-19-regulated genes. Using the TAIR website, we uploaded the sequences from Lewis et al. and obtained the 1000bp promoter sequences of the genes. With these sequences, we used FIMO to search for known AuxRE motifs found in the promoters of the potential genes where ARF-19 binds to turn on transcription. FIMO generated a list with targets that contain known ARF-19 motifs. Aspects of the gene that were considered before selection were the unique function of the gene, if it contains a potential motif that ARF-19 binds to, location of expression, if its expression is affected by an ARF-19 mutation, and the kinetics or timing of transcription abundance as compared to ARF-19. Gene AT5G64110 was originally selected as the target of interest, however, we were unable to successfully construct a plasmid with the genes promoter. Therefore, the focus of the study changed to the AT1G51950 (IAA18) gene.
PCR amplification of promoter and plasmid
The promoter of the desired gene was extracted from an Arabidopsis seedling. The seedling was grinding and treated with extraction buffer and isopropanol before centrifugation to isolate the DNA. Using the genomic DNA sequence of the AT5G64110 gene as a template, we designed the forward and reverse primers for polymerase chain reaction (PCR) to amplify the region. We aimed to design a primer that was 18-25 nucleotides long with GC content around 50%, had a melting temperature around 55° C, while avoiding repetitive and hairpin sequences. We added sequences from our expression vector, pGP6pIAA-Ubi-Venus-stop to both primers so that it will overlap with the vector sequence for successful assembly of the plasmid. PCR was then used to amplify the promoter regions of the target gene and to amplify the plasmid. PCR reactions consisted of our primers, the plant genomic DNA, water, and a Phusion Master Mix which was composed of buffer, Phusion polymerase, dTNPs, and MgCl2. The PCR products were verified through gel-electrophoresis. Gel-purification of the PCR products was then performed using a 0.7% agarose gel. The DNA band was excised and incubated with Binding Buffer at 60° C until the gel was completely dissolved. A silica-membrane column was then used to produce the purified DNA fragment.
Gibson cloning
The promoter sequence was inserted into the expression vector plasmid via Gibson Assembly. The assembly reaction consisted of three fragments including the promoter of the selected target gene, the Venus-EYFP reporter, Ura3, and ampicillin resistance to allow for selection of successfully transformed plasmids. The NEBuilding HiFi DNA assembly Master Mix was used to construct the plasmid with our insert.
Bacteria transformation
The bacteria E. coli was used to amplify the constructed plasmid. The plasmids from the assembly reaction were integrated into competent E. coli cells via heat shock at 42°C in a water bath. After the water bath, the vials were placed on ice for five minutes before being placed in the 37°C shaking incubator at 225 rpm and given S.O.C. rich media. After an hour, the transformation vials were spread on lysogeny broth (LB) agar plates containing ampicillin to select for positive transformation of our plasmids.
Plasmid DNA mini-prep
Plasmid DNA mini-preps were prepared for future integration into yeast for expression analysis. The bacterial cells were lysed by alkaline lysates and centrifuged at 13,000 rpm for ten minutes to isolate the plasmids from the other cell components. The supernatant was removed and centrifuged twice in a QIAprep 2.0 chromatography column. The DNA was eluted from the column using water, and centrifuged again to collect the pure plasmid DNA.
Restriction enzyme digestion and purification
It is necessary to first linearize the plasmid DNA for proper insertion into the budding yeast genome. We used NEBcutter V2.0 online program to produce the restriction map of our plasmid. Plasmid digestion was performed using PmeI restriction enzyme to linearize the plasmid. The complete reaction mL, which contained 10mL of the plasmid, 10mL Cut Smart Buffer, 2 mL of the PmeI enzyme, and 78 mL of water. We performed sequencing analysis using the BLAST online program to determine if the plasmid contains the correct insert of our desired gene. This program compares the similarity in the sequences between the genomic region of our gene and the mini-prepped plasmids sequences to indicate whether or not it was successfully inserted. It is important to note that this is the step of the procedure at which we changed our target gene because our primary selected gene promoter was not successfully inserted into the plasmid, as it was not identified by the BLAST program. A small sample of the digested product from another group with the IAA18 gene was then run through a 0.7% agarose gel to confirm a proper digestion had taken place.
The rest of the product was treated with 3M NaOAC and 100 mL isopropanol and stored at -20° C for 15 minutes. The solution was then centrifuged at 14,000 rpm for ten minutes the supernatant (non-DNA elements) was discarded. The DNA was treated with 100 mL 80% ethanol and centrifuged again for ten minutes to discard the supernatant. After this step, another short spin was performed to completely id the sample of supernatant, and the DNA was air dried. Lastly, 50 mL of water was added to the tube and carefully mixed.
Yeast transformation and PCR genotyping
The linearized plasmids from the E. coli amplification were transformed in the S. cerevisiae strain YKL381: matα LEU2: pGDP-AFB2 trp-1 ura3-1 his3-1. The cell mixture was heat shocked in a 42° C water bath for 15 minutes. The cell mixture was then centrifuged to form a cell pellet and remove the supernatant. Molecular water was then added to the cell pellet and suspended for placement on AA-Ura media agar plates to select for yeast cells with only our plasmids. The plates were incubated while inverted at 30° C until colonies appeared. The transformation of the cells was confirmed by zymolase digestion followed by PCR and gel electrophoresis on a 1.2% agarose gel. The positive colonies were spread on agar plates and saved.
Generation of yeast strain with target gene and ARF-19
The haploid cell matα leu2-3/LEU2: pGDP-AFB2 ura3-1/URA3:pGP6pIAA18-Ubi-Venus-stop trp-1 his3-1 was mated with the yeast haploid cell that contained the ARF-19 gene, matα his3-1/HIS3: Padh1-ARF-19 trp-1 ura3-1 leu2-3 to generate the matα/a leu2-3/LEU2: Pgdp-AFB2 ura3-1/URA3: Pgp6pIAA18-Ubi-Venus-stop his3-1/HIS3:Padh1-ARF-19 trp-1 strain. This strain now contains the desired gene promoter and ARF gene. Two other strains, 2320 matα/a leu2-3/LEU2:pGDP-AFB2 ura3-1/URA3: P3-2X-Ubi-Venus-stop his3-1 /HIS3: pADH1-ARF-19 trp-1 (2320) and 2466 matα/a leu2-3/LEU2:pGDP-AFB2 ura3-1/URA3: P3-2X-Ubi-Venus-stop his3-1 /HIS3: pADH1-ARF-19-H170A trp-1 (2466) were used as controls strains. The 2320 strain contained the wilt type ARF-19 gene serving as the positive control while the 2466 strain contained an ARF-19 gene with a mutation in DNA binding domain 1 served as the negative control.
RNA preparation and cDNA synthesis
RNA from the cells of all three strains of yeast was purified using the MasterPure RNA purification kit and protocol. The purity of the RNA samples was measured by the ratio of absorbance at 260nm and 280nm (260:280). From the pure RNA samples, complimentary DNA was produced for PCR to evaluate expression levels of our gene. All three samples had a negative control without reverse transcriptase to monitor for genomic contamination.
qRT-PCR, Confocal Microscopy, Plate Reader
Three methods were utilized to detect and quantify the expression of the selected gene’s promoter. All three Venus-reporter samples and a negative control of actin were run through the qRT-PCR reaction. After qRT-PCR, the products were run a 2% agarose gel to evaluate expression. A confocal microscope was used to take high resolution pictures of the cells from all three yeast strains to analyze the presence of yellow fluorescence that would be produced from the Venus-EYFP promoter. Two plate reader assays were run to measure the optical density and fluorescence signals of all three Venus-reporter samples: the IAA18 promoter strain, 2320, and 2466.
Statistical analysis
An ANOVA statistical test was applied to the data from the fluorescent measurement from the plate reader experiment to determine statistical significance between the three samples. P-values below the critical value of 0.05 were considered statistically significant.
Results
AT5G64110 gene as a potential target
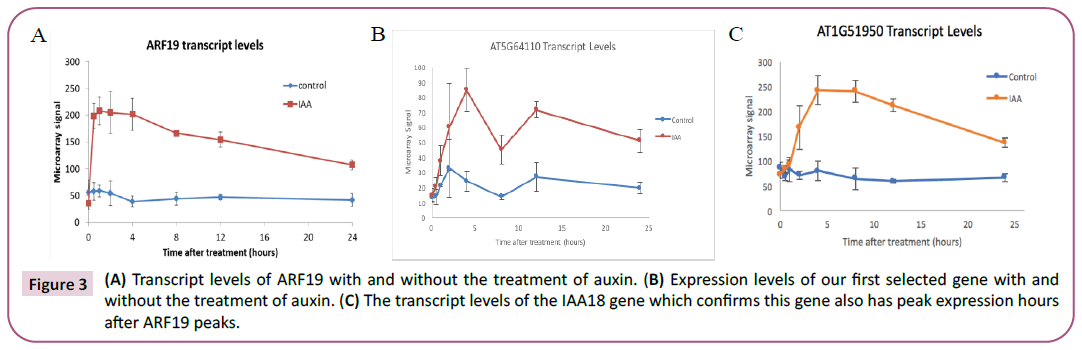
The AT5G64110 gene is a good target for this study as it met the criteria for many of the aspects that we considered. The gene contained a known ARF-19 motif, AuxRE from ARF5 was the identified motif in our gene. When analyzing the RNA-seq. data from Lewis et al. Compared to its expression in the plant with wild type ARF-19, in a mutant version, the fold change was -1.46, signifying there was a significant decrease in expression when ARF-19 is mutated and dysfunctional (p=9.44, E-03) (Table 1) [4]. It is localized in the extracellular region of both roots and shoots in the plant. The timing of the peak of transcription of the gene occurs hours after auxin treatment and ARF-19 peak expression levels (Figure 3).
Figure 3: (A) Transcript levels of ARF19 with and without the treatment of auxin. (B) Expression levels of our first selected gene with and
without the treatment of auxin. (C) The transcript levels of the IAA18 gene which confirms this gene also has peak expression hours
after ARF19 peaks.
Production of promoter and plasmid
The forward primer that we used was ggtcgctcttattgaccacacctccgccagCAGAGAAAATGGCCGACAATC, and the reverse primer was accggtcaaagtcttgacgaaaatctgcatCGGTAAGAGAACGTAGCAAGG. Using our designed forward and reverse primers, we successfully amplified the promote region of our gene using PCR (Figure 4). The Gibson Assembly reaction was confirmed to have successfully inserted the gene promoter and other two fragments through gel electrophoresis (Figure 5).
Figure 4: Gel electrophoresis of the AT5G64110 promoter region
from PCR. The red oval indicates the DNA band that was
excised and purified using gel-purification.
Figure 5: Gel electrophoresis of the Gibson Assembly products, which shows that our group successfully constructed the plasmid as indicated by one strong band (indicated by red arrow).
PmeI restriction digestion and yeast transformation
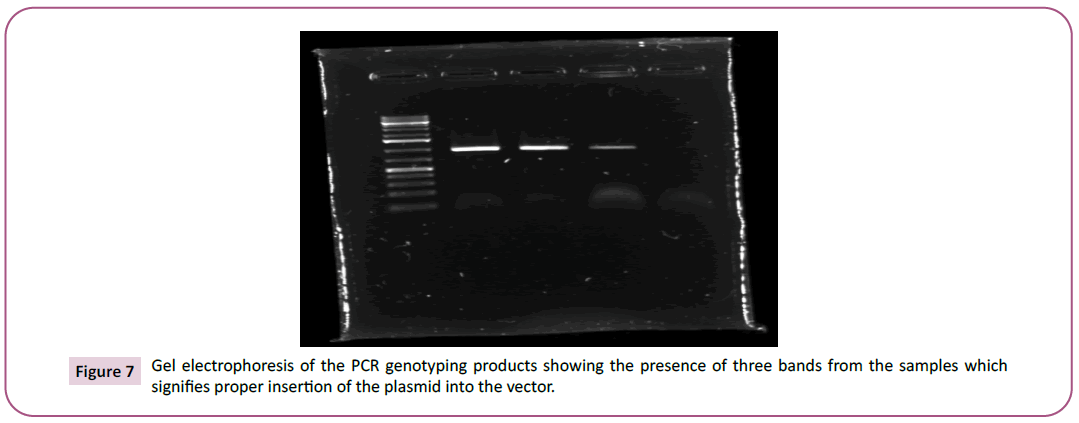
As indicated in Figure 5), we believed to have successfully insert our gene’s promoter into the plasmid. However, the BLAST program disproved this, and the promoter region was not actually inserted in the plasmid. Here, we switched to the IAA18 gene, and used the plasmid constructed by another group using the same method to perform the digestion. PmeI successfully digested the plasmid as indicated by the two bands on the gel in (Figure 6). The linearized plasmid from the E. coli was then successfully transformed into the budding yeast, as confirmed by gel electrophoresis of the PCR genotyping (Figure 7). The visualization of three bands on the gel indicate that the vector was successful inserted into the yeast genome (Figure 7).
Figure 6: Gel electrophoresis of the digested plasmids by PmeI. Our groups products are highlighted in the red oval.
Figure
Figure 7: Gel electrophoresis of the PCR genotyping products showing the presence of three bands from the samples which
signifies proper insertion of the plasmid into the vector.
Observation and quantification of IAA18 gene expression
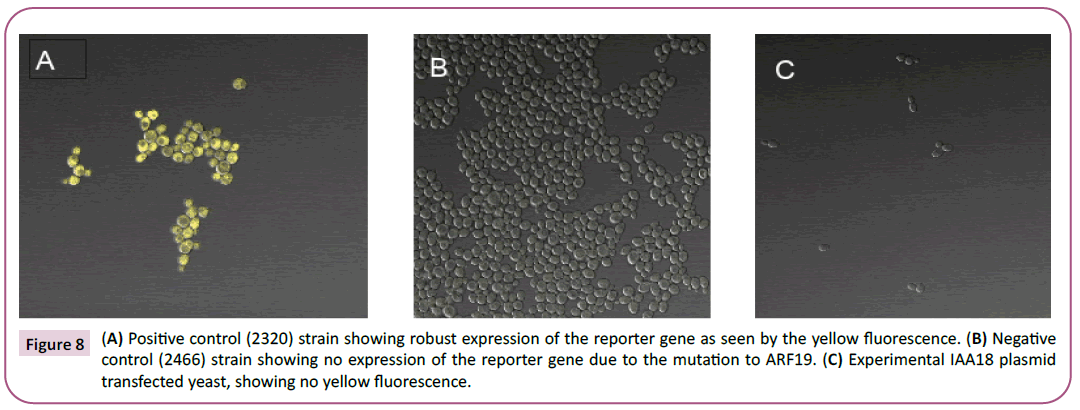
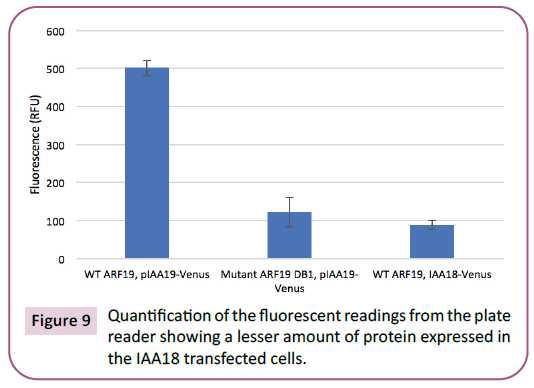
Overall, our gene promoter did not drive the expression of the EYFP-Venus reporter gene in yeast. All three of the methods used to analyze the expression correspond to each other and indicate that expression did not occur. Thus, ARF-19 did not bind to the promoter and activate transcription. Using the confocal microscope, we observed no yellow fluorescence, as it reflected the appearance of the negative control (Figure 8). The fluorescence signals obtained from the plate reader support the result that there was no expression. The average fluorescence measured of the positive control, negative control, and our gene’s strain was 502, 122 and 88RFU, respectively (Figure 9). The ANOVA test confirmed that there was a statistical difference between our gene’s expression and the positive control (p =2.224E-6). The qRT-PCR of the three samples was not successful, as no bands appeared on the gel (Figure 10).
Figure 8: (A) Positive control (2320) strain showing robust expression of the reporter gene as seen by the yellow fluorescence. (B) Negative
control (2466) strain showing no expression of the reporter gene due to the mutation to ARF19. (C) Experimental IAA18 plasmid
transfected yeast, showing no yellow fluorescence.
Figure 9: Quantification of the fluorescent readings from the plate
reader showing a lesser amount of protein expressed in
the IAA18 transfected cells.
Figure 10: The gel electrophoresis of the qRT-PCR products
showing no visualization of bands in the positive Venus
control, negative Venus control, IAA18 experimental
group, or the actin negative control.
Discussion
The AT5G64110 gene was originally selected as our group was interested in its function as a peroxidase superfamily protein involved in hydrogen peroxide catabolism which can damage DNA and induce oxidative stress on the organism. The examination of this gene was disrupted during the construction of the plasmid vector to be, as the promoter gene was not identified in the sequence of the plasmid. Therefore, the expression analysis was performed on the IAA18 gene.
It is clear that IAA18 is not a direct downstream target of ARF-19, and thus, further studies can be directed towards other potential targets of ARF-19 to uncover other genes involved in the ARF-19-auxin mediated response pathway. Our confocal images and plate reader data confirm this result. The confocal image shows that no expression takes place, as the amount of YFP produced from the IAA18 strain reflects the negative control strain. The plate reader fluorescent experiment confirms that there is little IAA18 protein in the yeast.
Our qRT-PCR reaction was inconclusive (Figure 10). It is possible that the RNA was contaminated, the cDNA synthesis did not work, or the PCR reaction just did not run correctly. The expression of IAA18 may occur independently of ARF-19 activity, or could be enhanced through an intermediary gene of ARF-19. Further research on the IAA18 transcription factor could provide more information on plant growth and development and other plant processes that occur outside of the auxin-ARF-19 pathway. IAA18 has been found in the apical domain of globular stage embryos, and an iaa18-1 mutant has been seen to cause defects in cotyledon location and structure [5]. Further analysis of other mutations of these gene can help researchers understand its function and interaction with other potential ARFs in plants. A previous study actually confirmed that this same gain of function mutation, iaa18-1, blocks lateral root formation in crane-1 and crane-2 Arabidopsis mutants [6]. The same study also reported that IAA18 proteins interact with ARF-7 and ARF-19 in yeast, in that it works with IAA14 to as a repressor of these to ARFs [6]. This could suggest that the IAA18 gene is expressed by the function of a separate ARF, and the protein then inhibits ARF-19 from functioning. Our results would support this finding in that ARF-19 does not activate IAA18 transcription.
Conclusion
This study makes it clear that future research is required to identify target genes of ARF-19 in the auxin-controlled physiological response. However, this study did confirm that yeast is a model organism to assess the transcription factors ability to bind to and promote expression of a desired promoter.
References
- Pierre-Jerome E, Moss BL, Lanctot A, Hageman A, Nemhauser JL (2016) Functional analysis of molecular interactions in synthetic auxin response circuits. Proc Natl Acad Sci 113: 11354–11359.
- Korasick DA (2014) Molecular basis for Auxin Response Factor (ARF) protein interaction and the control of auxin response repression. Proc Natl Acad Sci. 111: 5427–5432.
- Yoko O (2007) ARF-7 and ARF-19 Regulate Lateral Root Formation via Direct Activation of LBD/ASL Genes in Arabidopsis. The Plant Cell 19: 118–130.
- Lewis DR (2013) A Kinetic Analysis of the Auxin Transcriptome Reveals Cell Wall Remodeling Proteins That Modulate Lateral Root Development in Arabidopsis. The Plant Cell 25: 3329–3346.
- Ploense SE, Wu MF, Nagpal P, Reed JW (2009) A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136: 1509–1517.
- Takeo U (2008) Domain II Mutations in CRANE/IAA18 Suppress Lateral Root Formation and Affect Shoot Development in Arabidopsis Thaliana. Plant and Cell Physiology 49: 1025–1038.