Keywords
Human immunodeficiency virus; CD4; Anti-CD4 antibody; Infection blocking
Introduction
Human CD4 is a single-chain, trans-membrane glycoprotein belonging to the immunoglobulin superfamily that is mainly expressed on T lymphocytes, B lymphocytes and thymocytes. CD4 plays a vital role in T cell maturation and signal transduction, acts as an anchoring target in comprehensive antigen responses, and is indispensable in adaptive immunity.
Human CD4 has a relative molecular weight of 55kD. The local genome size of 6.77~6.80 Mb, comprising 10 exons, is found between human chromosome 12P12 and 12pter [1]. The CD4 molecule precursor consists of 458 amino acids (aa), which includes a 25-aasignal peptide, a 371-aa extracellular region, a 24-aa trans-membrane region and a 38-aa cytoplasmic region. Four immunoglobulin-like domains are situated extracellularly: D1 (1~98 aa), D2 (99~178 aa), D3 (179~291 aa) and D4 (292~371 aa), from N- to C-terminal, respectively [2]. The crystal structure of CD4 (Figure 1) shows that D1 and D3 resemble Immunoglobulin Variable (IgV) domains, whereas D2 and D4 resemble Immunoglobulin Constant (IgC) domains [3]. The D1 core domain consists of two β-sheets formed by nine β-strands that are linked by a disulfide bond bridge. D2 connects with D1 through its hydrophobic interface, as does D3 with D4. Additionally, residues 95 to 100 and 174 to 179 are similarly hydrophobic, indicating that D2and D3 are joined mainly through hydrophobic interactions [4]. D3 contains no disulfide bonds, with its two β-sheets separated from each other. D4, structurally resembling D2, is widely believed to activate T cells and CD4 function through the dimerization of CD4 molecules [3,5]. The transmembrane region is hydrophobic whereas the intracellular region comprises three serine residues (S408, S415 and S431) that are phosphorylated to mediate signal transduction. These serine residues connect directly with the Src Tyrosine Kinase (TK) family member P56lck, which can increase the level of P56lck tyrosine phosphorylation and regulate signal transduction [6].
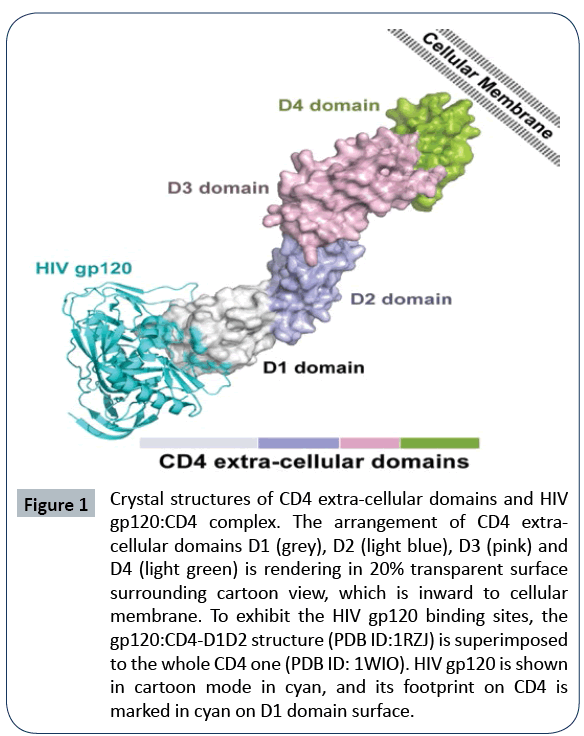
Figure 1: Crystal structures of CD4 extra-cellular domains and HIV
gp120:CD4 complex. The arrangement of CD4 extracellular
domains D1 (grey), D2 (light blue), D3 (pink) and
D4 (light green) is rendering in 20% transparent surface
surrounding cartoon view, which is inward to cellular
membrane. To exhibit the HIV gp120 binding sites, the
gp120:CD4-D1D2 structure (PDB ID:1RZJ) is superimposed
to the whole CD4 one (PDB ID: 1WIO). HIV gp120 is shown
in cartoon mode in cyan, and its footprint on CD4 is
marked in cyan on D1 domain surface.
Functionally, CD4 interacts directly with major Histocompatibility Complex (MHC) class II molecules on the surface of antigen presenting cells and helps recruit P56lckto facilitate the activation of helper T cells, thus modulating the adaptive immune response [7]. The interaction of CD4 with MHC II was defined by structural studies of a complex containing CD4D1 and D2with the murine I-Ak class II MHC molecule and bound peptide (pMHCII) [8]. The complex structure revealed that the CD4 N-terminal IgVlike domain directly reaches into the two membrane-proximal domains of the pMHCII molecule. The C-terminus of CD4 points away from pMHCII, indicating that the pMHCII molecule mainly interacts with the D1 domain. In contrast, the D2 domain makes no contact with the pMHCII molecule [8].
CD4 is noted to be the primary receptor for Human Immunodeficiency Virus (HIV)-1 infection. CD4 binds to gp120 to interrupt the membrane fusion process and initiate infection. Gp120 is one of the two domains of the maturing HIV-1 membrane envelope glycoprotein precursor gp160; the other is gp41. The CD4–gp120 interaction constitutes the first step in HIV-1 attachment, which is followed by gp120 binding to a second cellular receptor, either Chemokine Receptor-5 (CCR5) or CX Chemokine Receptor-4 (CXCR4) [9]. This secondary binding allows the gp41(fusion peptide) molecule of HIV-1 to insert into the host cell membrane, eventually mediating membrane fusion of the virus with the host [10,11]. CD4 thus has a key role in the initiation of HIV-1 infection. Comparing bound and unbound crystal structures of gp120 withCD4 shows that a “bridging sheet”—a four-stranded β-sheet formed by two β-hairpins—fixes the relative orientations of the two closely associated “inner” and “outer” domains of the gp120 core duringCD4 binding [12]. The CD4 D1 domain interacts with these inner and outer domains as well as the bridging sheet, which leadsto the rearrangements of the gp120 inner domain [13]. Furthermore, with additional interactions with the gp120 V3 variable loop, the bridging sheet exposes the co-receptor binding site [14,15].
Numerous anti-CD4 antibodies have been described over the past decades: those recognizing the D1 domain (e.g. Q4120, 6H10, 2D5, and 2F2), the D2 domain (e.g., mAb Mu5A8, Leu3A, OKT4A, F91-55, and M-T441) and the D3 and D4 domains (e.g.,mAb OKT4 and L120). All of these CD4 antibodies display various properties, and these have triggered much research on the efficacy of CD4 antibodies in neutralizing HIV-1 infection. In the next section, we will primarily discuss two representative antibodies, mAb 15A7 and Ibalizumab, as examples of D1- and D2-specific antibodies, respectively.
D1-Specific Antibodies
mAb 15A7
In terms of its neutralization effect in vitro, mAb15A7 demonstrates the most optimal targeting of the CD4 D1 domain. It can neutralize HIV-1 isolates covering the B, C, D, D/E subtypes at an IC50 of 0.06 μg/ml to 0.37 μg/ml [16,17]. Epitope mapping of the 15A7 binding site via alanine scanning mutagenesis showed that 15A7 recognizes a conformational epitope. Using knowledge-based molecular docking, the binding residues on the CD4 D1 domain were mapped as F26, H27, K29, K35, Q40, S42, F43, L44, T45, K46, N52, D56, S57, R59, S60, W62 and K72. Binding is reported to be achieved via the hydrophobic loops within the variable regions of 15A7, with slight statistical differences in affinity in the presence and absence of HIV-1, as determined by cell-based CD4 binding assay. Presumably, the 15A7 binding site partially overlaps with the gp120 binding site. A cartoon model of the 15A7-D1D2-gp120 interaction was adapted to reveal this structural interaction (Figure 2). An inhibition assay further showed that the bivalent full-length mAb 15A7 or its F(ab’)2 completely blocks gp120 binding, which is 20 times stronger than its Fab form. As to its CD4 attachment ability, 15A7 can almost completely bind to CD4- expressing cells, such as TZM-Bl, U87.CD4.CCR5, H9 and MT4; however, only 51% and 38% binding is noted between 15A7 and CD4 on PBMC and U87.CD4.CXCR4 cells, respectively. These results strongly suggest that 15A7 targets a particular portion of CD4 in susceptible cells.
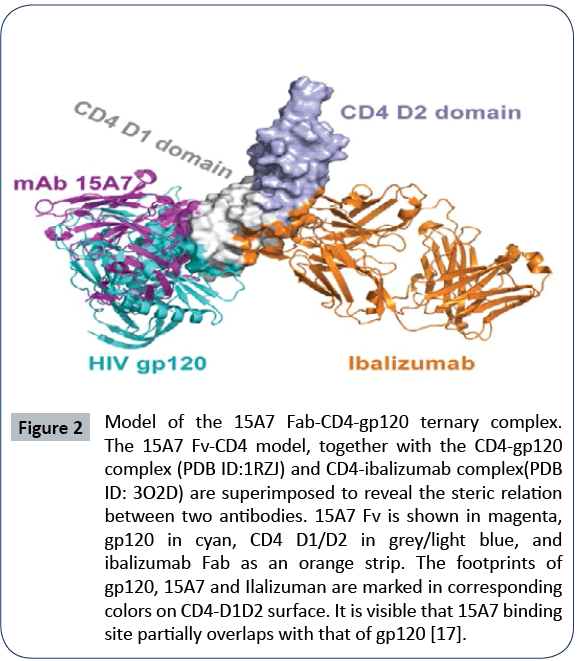
Figure 2: Model of the 15A7 Fab-CD4-gp120 ternary complex.
The 15A7 Fv-CD4 model, together with the CD4-gp120
complex (PDB ID:1RZJ) and CD4-ibalizumab complex(PDB
ID: 3O2D) are superimposed to reveal the steric relation
between two antibodies. 15A7 Fv is shown in magenta,
gp120 in cyan, CD4 D1/D2 in grey/light blue, and
ibalizumab Fab as an orange strip. The footprints of
gp120, 15A7 and Ilalizuman are marked in corresponding
colors on CD4-D1D2 surface. It is visible that 15A7 binding
site partially overlaps with that of gp120 [17].
Further verification of the residues identified through alanine scanning has been conducted by examining CD4 fragments in their interactions with 15A7. Both ELISA assay and 293FT-based flow cytometry have shown that 15A7 can bind the D1D2 and the D1 fragments of CD4 but not D3 or D4 fragments. Reciprocally, the CD4 fragments (D1 and D1D2) can block attachment to the cell and cause neutralization of the mAb 15A7. The mAb 15A7 also demonstrates a mild binding affinity to other D2 antibodies, which indicates humanized and affinity maturation should be implemented in future applications. Moreover, 15A7 might exert its inhibition through a post-attachment mechanism, which is similar to that seen with mAb Q4120 [18]. 15A7 can inhibit 80% of HIV-1 entry at from 30 to 120 minutes post-infection, and this inhibitory effect is dependent on the antibody concentration. In general, however, 15A7 may not activate the P56lck-mediated signaling pathway of CD4 [6].
During the process of HIV infection, both CD4 and gp120 vary in their respective conformations, and this knowledge, in addition to the broad HIV-1 neutralization capacity of 15A7, might help to improve the development of inhibitors for HIV-1 treatment.
Other d1 domain antibodies
D1 domain antibodies competitively inhibit HIV-1 gp120 anchoring to CD4 [19]. Although these antibodies display high affinity to CD4, they show differential neutralization capacities, and these limitations need to be addressed before any of these antibodies can be considered for therapy [20]. Murine mAb 2D5 binds both human and rhesus CD4, similar to Leu3A and its homologous hybridoma antibody 2F2: yet, 2D5 displays better affinity to rhesus CD4, and Leu3A shows better sensitivity to human CD4. When comparing the neutralization potency of 2D5, Leu3A and 2F2 to isolate HIV-1 SF162, Leu3A shows a better inhibition (IC50 ~0.01 μg/ml), whereas 2D5 (~0.08 μg/ml)and 2F2 (~0.1 μg/ ml). It is worth noting that 2D5 and 2F2 display obviously better neutralization against SHIV SF163P3 than the anti-CD4 binding site mAb VRC01 (about 15-fold above) [21]. Notably, although CD4 mAb 2D5 and 2F2 display greater potency than the Env mAb, they have been found to confer little in vivo protective efficacy [21].
D2-specific antibodies: Ibalizumab:
Ibalizumab (TMB-355; formerly TNX-355 or Hu5A8), is a monoclonal antibody (mAb) derived from the precursor, Mu5A8. Mu5A8 recognizes the D2 domain through residues 121 to 134, as defined by alanine scanning in chimeras of murine and human CD4 [20,22]. Earlier reports showed that Mu5A8 acts synergistically with an anti-gp120 V3 loop antibody, NEA-9205, to inhibit the virus and cell membrane fusion [23]. Moreover, Mu5A8 is efficient in HIV-1 infection in highly diluted serum samples from patients, even in very low antibody concentrations [24].
The humanization of Mu5A8 was conducted by TanoxInc. (Houston, TX). This saw there placement of the Mu5A8 CDR region with human IgG4, and mutations to amino acids in the FR region, which resulted in 95% humanization of ibalizumab and its a remarkably long half-life [9]. The epitope of ibalizumab sits mainly on the D2domain of CD4—residues P121, P122, and Q163—with contribution from residue E77 and probably S79 and L96 of the D1 domain [25]. This epitope does not interfere with gp120 binding to CD4. Instead, it appears to exert its inhibition effect by post-binding conformation effects, which prevent gp120 from approaching the CCR5 or CXCR4 co-receptors [26,27]. The neutralization ability of ibalizumab to HIV-1 isolates demonstrates an IC50 of 0.01 to 0.13 μg/ml for C subtype isolates; IC50 of 0.02 to 2.20 μg/ml for B subtype isolates, IC50 of 0.02 to 0.1 μg/ ml for A subtype isolates; and the isolates Q769.22 and SF162. LS show over 100 μg/ml [8]. Clinically, amongst a large panel of 118 relevant HIV-1 pseudoviruses, ibalizumab neutralizes 92% of viruses with half-infection inhibition, and 47% of viruses with 90% infection inhibition. An evaluation in vivo indicates that ibalizumab also shows an effective ability to decrease plasma viral loads and increase CD4+ cell counts in HIV-1-infected patients and rhesus monkeys infected with simian immunodeficiency virus [28].
Molecular Dynamic (MD) stimulations were performed to map the interaction sites on ibalizumab using the ibalizumab–CD4 receptor complex (PDB ID: 3O2D). Five key residues were noted to play an essential role in the ibalizumab–CD4 interaction: Y50(HCDR2), Y53(HCDR3), D58(HCDR2), E95(HCDR3), and R95(LCDR3) [29].
Other D2 domain antibodies
M-T441 is another D2-specific antibody with neutralizing activity. The epitope of M-T441 covers residues V123, F124, G125, G138, S139 and L140. The KD(M) of M-T441 is nearly 10-folder weaker than that of ibalizumab, and its neutralization ability is reported to be positive against the isolates of JRCSF, MDR-1a, MDR-5a, and WT-1a (with the IC50 ranging from 0.2 μg/ml to 7 μg/ml) [26]. Thus, its neutralization potency is still far lower than that of ibalizumab. One possible explanation for this result is that its epitope is a little lower down on D2. Another antibody, OKT4A, displays preferable HIV-1 infection blocking ability but it is extremely immunosuppressive because it interferes with the physiological function and signaling pathway in the cell [30].
Further Optimization of Anti-CD4 Antibodies
By most standards, ibalizumab is the most wholly effective of the antibodies identified to date. However, some HIV-1 strains bear mutations that are resistant to ibalizumab, such as that found following the loss of an N-linked glycan from the V5 loop of Env gp120 [31].This resistance was resolved by introducing, under denaturing conditions, potential N-linked glycosylation sites to residue 52 of the ibalizumab L-chain variable region, which residesclose to the gp120 V5 loop. This new, optimized antibody, LM52, showed remarkably improved neutralization potency and breadth as compared with ibalizumab. Indeed, LM52 has been shown to neutralize a panel of 118 diverse HIV-1 viral strains covering 11 clades, and also displaysan IC50 value under 0.1 μg/ml for all tested isolates as compared with the 75% of isolates achieved for ibalizumab [31]. These findings indicate that the strategic placement of aglycan in the antibody variable region could improve its functional activity. Indeed, further relevant studies have since shown that the N-linked carbohydrate replacement strategy in the variable region leads to improved solubility [32].
Ibalizumab-based, Bi-Specific Broadly Neutralizing Antibodies (BibNAb) have also helped to reveal certain aspects of HIV-1 infection prevention. PG9-ibalizumab (PG9-iMab) and PG16-iMab were created by fusing the scFv of PG9 or PG16, respectively, to the N-terminus of the H chain of ibalizumab through a flexible linker [33,34]. These reconstructed BibNAbs demonstrate potent and wide neutralization against 100% of the tested viruses (118/118), including viruses that demonstrated resistance to the parental antibodies [35]. These BibNAbs also showed significant potency at low (picomolar) concentrations, demonstrating their clear efficacy. Considering that bi-specific antibodies have been successful in the clinic, the wide and potent neutralizing activities of PG9-iMab and PG16-iMab suggests its suitability as a candidate immunization therapy for HIV-1 treatment [36].
Another antibody, iMabm36, consists of ibalizumab and two copies of the single-domain antibody m36. The epitope of m36 is on a highly conserved CD4-induced region; yet, the optimized activity of iMabm36 relies to some extent on the binding sensitivity of m36 [37]. Obviously, the improved antiviral activity of iMabm36 is attributed to the dual mechanism of its parental antibodies. But it displays enhanced antiviral activity compared with either parental antibody alone, or in combination (at the coadministration ratio of Imab:m36=1:2, 1:10) [37].
Overall, although ibalizumab and its recombined antibodies show promise in HIV-1 infection blocking, the characterization and research of CD4 and its antibodies still remains a great challenge.
Concluding Remarks and Thoughts for the Future
After decades of research, several broad-neutralizing mAbs against HIV-1 have been developed to eradicate HIV-1. The current suite of available neutralization antibodies act by blocking HIV-1 entry into the host cell. As we detail, CD4 antibodies display obvious and characteristic features that block HIV-1 infection. CD4 antibodies interfere with the HIV-1 infection process by more than one mechanism but have different effects depending on when the antibody is delivered over the course of HIV-1 infection (as is described in 15A7 post-attachment-neutralization): (1) When introduced to CD4 ahead of infection, CD4 antibodies sterically prevent HIV-1 from attachment to host cell; (2) When HIV-1 has already attached the host cell, CD4 antibodies (for example, 15A7 and Q4120) may retain their post-attachment-neutralization ability and interfere with membrane fusion; (3) When HIV-1 infection is ongoing, CD4 antibodies inhibit membrane fusion by competitively binding to CD4 (D1-specific antibodies) or by sterically blocking co-receptor binding (ibalizumab and Mu5A8).
High-throughput, next-generation sequencing of B cells and single-cell PCR-amplification assays have helped to identify the effective CD4-binding-site antibodies and traditional methods that screen for CD4-specific antibodies can also be useful in generating effective antibodies for HIV-1 treatment [38,39]. An increasing number of reports suggest a significant synergistic effect of antibodies targeting HIV-1 Env and the host cell [40]. Theoretically, the combination of potent Env antibodies and CD4 antibodies could be used to suppress viral binding activity and protect host cells from attachment. Indeed, PG9-iMab and PG16-iMab have demonstrated this to be a promising option. Through similar mechanisms, 15A7 optimization may feasibly provide a bispecific antibody solution [41]. Notably, 15A7 differs from any other reported CD4 antibody in terms of its outstanding neutralization capacity and D1-specificity.
In general, CD4 antibodies are unlikely to suffer from immune evasion mechanisms in the human immune system, and this establishes their distinct superiority over other HIV-specific antibodies. Their non-immunosuppressive, potent and widereaching effects against HIV-1 infection, concomitant with their high affinity to target CD4 domains, could make CD4 antibodies the therapeutic choice for HIV-1 treatment in the future.
References
- Ansari-Lari MA, Muzny DM, Lu J, Lu F, Lilley CE, et al. (1996) A gene-rich cluster between the CD4 and triosephosphate isomerase genes at human chromosome 12p13. Gnome Res 29: 293-298.
- Sharma D, Bella MM, Charkraborty K, Kumaran S, Jeqanathan S, et al. (2005) Protein minimization of the gp120 binding region of human CD4.Biochemistry 44: 16192-16202.
- Wu H, Kwong PD, Hendrickson WA (1997) Dimeric association and segmental variability in the structure of human CD4. Nature 387: 527-530.
- Harrison SC, Wang J, Yan Y, Garrett T, Liu J, et al. (1992) Structure and interactions of CD4.Cold Spring Harb Symp Wuant Biol 57: 541-548.
- Moldovan MC, Sabbaqh L, Breton G Sekaly RP, Krummel MF(2006) Triggering of T Cell Activation via CD4 Dimers. The Journal of Immunology 176: 5438-5445.
- Brenner B, Gulbins E, Busch GL, Walzoq B, Lanq F, et al.(1996) L-selectin activates the Ras pathway via the tyrosine kinase p56lck. Proc Natl Acad Sci USA 93: 25376-25381.
- Doyl C, Strominger JL(2010) Interaction between CD4 and class II MHC molecules mediates cell adhesion. J Immuno 184: 5935-5938.
- Freeman MM, Seaman MS, Rits-Volloch S, Hong X, Ho DD, et al.(2010) Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody. Structure 18: 1632-1641.
- Bruno CJ, Jacobson M(2010) Ibalizumab: an anti-CD4 monoclonal antibody for the treatment of HIV-1 infection. J Antimicrob Chemother 65: 1839-1841.
- Kuritzkes, Daniel R (2009) HIV-1 entry inhibitors: an overview. Curr Opin HIV AIDS 4: 82-87.
- Briz V, E Poveda, V Soriano(2006) HIV entry inhibitors: mechanisms of action and resistance pathways. J Antimicrob Chemother 57: 619-627.
- Chen B, Vogan EM, Gonq H, Skehel JJ, Wiley DC, et al. (2005) Structure of an unliganded simian immunodeficiency virus gp120 core.Nature 433: 834-841.
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, et al.(1998) Structure of an HIV gp120 core envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody.Nature 393: 648-659.
- Huang CC, Lam SN, Acharya P, Tang M, Xiang CH, et al.(2007) Structures of the CCR5 N terminus and of a tyrosin-sulfated antibody with HIV-1 gp120 and CD4. Science 317: 1930-1934.
- Harrison CC (2008) Viral membrane fusion. Nat Struct Mol Biol 15: 690-698.
- Burastero SE, Figini M, Frigerio B, Lusso P, et al.(2009) Protective versus pathogenic anti-CD4 immunity: insights from the study of natural resistance to HIV infection. Journal of Translational Medicine 7: 101.
- Hou W, Liu JY, Yu H, Qi JL, Zhang ZQ, et al.(2015) Molecular insights into the inhibition of HIV-1 infection using a CD4 domain-1-specific monoclonal antibody. Antiviral Research 122: 101-111.
- Mclnerney TL, Dimmock NJ (2001) Postattachment neutralization of a primary strain of HIV type 1 in periheral blood mononuclear cells is mediated by CD4-specific antibodies but not by a glycoprotein 120-specific antibody that gives potent standard neutralization.AIDS Res Hum Retroviruses 17: 1645-1654.
- Endres MJ, Clapham PR, Marsh M, Ahuja M, Turner JD, et al.(1996) CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 9: 745-756.
- Moore JP, Sattentau QJ, Klasse PJ, Burkly LC (1992) A monoclonal antibody to CD4 domain 2 blocks soluble CD4-induced conformational changes in the envelope glycoprotein of human immunodeficiency virus type 1(HIV-1) and HIV-1 infection of CD4+ cells. J Virol 66: 4784-4793.
- Pegu, A, Yang ZY, Boyington JC, WU L, Ko SY, et al.(2014) Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Transl Med 6: 243-258.
- Chibisa GE, Takahashi Y, Lichtenfeld J, Tanaka R, Yoshida A, et al.(2001) Functional CD4 Tcell after intercellular molecular transfer of 0X40 ligand. J Immuno 167: 875-883.
- Laal S, Murey N, Blumenthal R, Dimitrov DS (1995) Syenergistic inhibition of human immunodeficiency virus type 1 envelope glycoprotein-mediated cell fusion and infection by an antibody to CD4 domain 2 in combination with anti-gp120 antibodies. J Virol 69: 4267-4273.
- Burkly L, Burda S, Gorny MK, Karwowska S, Buchbinder A, et al.(1994) Syneric neutralization of huaman immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J Virol 68: 4001-4008.
- Li L, Shi X, Lu Q, Zhang S, Wang X, et al. (2013) Role of human CD4 D1D2 domain in HIV-1 infection. Immunol Invest 42: 106-121.
- Song, R, Franco D, Kao CY, Yu F, Huang Y, et al. (2010) Epitope mapping of ibalizumab, a humanized anti-CD4 monoclonal antibody with anti-HIV-1 activity in infected patients. J Virol 84: 6935-6942.
- Reimann KA, Khunkhun R, Lin W, Gordon W, Fung M (2002) A humanized, nondepleting anti-CD4 antibody that blocks virus entry inhibits virus replication in rhesus monkeys chronically infected with simian immunodeficiency virus. AIDS Res Hum Retroviruses 18: 747-755.
- Jacobson JM, Kuritzkews DR, Godofsky E, Dejesus E, Larson JA, et al. (2009) Safety, pharmacokinetics, and antiretroviral activity of multiple doses of ibalizumab (formerly TNX-355), an anti-CD4 monoclonal antibody, in human immunodeficiency virus type 1-infected adults. Antimicrob Agents Chemother 53: 450-457.
- Su ZY (2014) Ibalizumab-human CD4 receptor interaction: computational alaine scanning molecular dynamics studies. Curr Comput Aided Drug Des 10: 217-225.
- Pulitto R, Roberts VA, Adair JR, Rothermel AL, Collins AM, et al. (1996) Humanization and molecular modeling of the anti-CD4 monoclonal antibody, OKT4A. J Immuno 156: 2840-2850.
- Song, R, Oren DA, Franco D, Seaman MS, Ho DD (2013) Strategic addition of an N-linked glycan to a monoclonal antibody improves its HIV-1-neutralizing activity. Nat Biotechnol 31: 1047-1052.
- Pepinsky RB, Silvian L, Berkowitz SA, Farrington G, Lugovskov A, et al. (2010) Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis. Protein Sci 19: 954-966.
- Walker LM, Phogat SK, Chan PY, Wagner D, Phung P, et al. (2009) Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326: 285-289.
- Pace CS, Song R, Ochsenbauer C, Andrews CD, Franco D, et al. (2013) Bispecific antibodies directed to CD4 domain 2 and HIV envelope exhibit exceptional breadth and picomolar potency against HIV-1. Pro Natl Acad Sci USA 110: 2313-2348.
- Morgand M, Bouvin-pley M, Plantier JC, Moreau A, Alessandri E, et al. (2015) A V1V2 neutralizing epitope is conserved in divergent non-M groups of HIV-1. J Acquir Immune Defic Syndr. Inpress.
- Bargou R, Leo E, Zuqmasier G, Klinger M, Goebeler M, et al. (2008) Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science 321: 974-977.
- Sun MB, Pace CS, Yao X, Yu F, Padte NN, et al. (2014) Rational design and characterization of the novel, broad and potent bispecific HIV-1 neutralizing antibody iMabm36. J Acquir Immune Defic Syndr 66: 473-483.
- Lynch RM, Tran L, Louder MK, Schmodt, Cohen M, et al. (2012) The development of CD4 binding site antibodies during HIV-1 infection. J Virol 86: 7588-7595.
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, et al. (2001) Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 75: 10892-10905.
- Wu L, Zhou T, Yang ZY, Svehla K, O'Dell S, et al. (2009) Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol 83: 5077-5086.
- Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, et al. (2013) Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 155: 531-539.