- (2011) Volume 12, Issue 4
Xunhai Xu1, Alexios S Strimpakos2, Muhammad Wasif Saif1*
1Division of Hematology and Oncology, Columbia University Medical Center. New York, NY, USA
2Oncology Unit, Third Department of Medicine, Sotiria General Hospital. Athens, Greece
Appropriate identification and validation of biomarkers as well as pharmacogenetics are important in formulating patient-oriented, individualized chemotherapy or biological therapy in cancer patients. These markers can be especially valuable in pancreatic cancer, where high mortality and complex disease biology are frequently encountered. Recently, several advances have been made to further our knowledge in this specific area of pancreatic cancer. In the 2011 American Society of Clinical Oncology (ASCO) Annual Meeting, researchers have presented several interesting results in biomarkers development: the identifications of 9 single nucleotide polymorphisms (SNPs) that is associated with positive efficacy of gemcitabine (Abstract #4022); the introduction of circulating tumor cells as a prognostic markers in pancreatic adenocarcinoma (Abstract #e14657); the re-affirmation of plasma cytidine deaminase (CDA) as a positive predictive markers for gemcitabine efficacy, as well as the postulations that CDA*3 as a potential genotype marker to predict gemcitabine responses (Abstract #e14645); and finally the retrospective tumor tissues analysis in the Arbeitsgemeinschaft Internistische Onkologie (AIO) trial in an attempt for epidermal growth factor receptor (EGFR) pathway biomarker identifications (Abstract #4047)
Biological Markers; Biological Therapy; gemcitabine; Neoplastic Cells, Circulating; Pancreatic Neoplasms; Pharmacogenetics
AIO: Arbeitsgemeinschaft Internistische Onkologie; CDA: cytidine deaminase; CI: confidence interval; SNP: single nucleotide polymorphism
Pancreatic adenocarcinoma is one the most aggressive cancers worldwide. Current standard chemotherapy for pancreatic cancers remains to be gemcitabine-based. With the introduction of targeted therapy, several new platforms of therapies such as anti-epidermal growth factor receptor (EGFR) agents are current being evaluated. At same time, many attempts have also been made to incorporate effective target-oriented biomarkers and pharmacogenetics in formulating individualized cancer treatment.
Biomarkers are either prognostic or predictive. Prognostic biomarkers are intrinsic indicators for tumor’s aggressiveness and patients’ final clinical outcome, regardless of the therapy received. Their clinical relevance is significant as they allow for better risk stratifications as well as rapid assessment of likelihood of disease progression or recurrence. On the other hand, predictive markers are parameters used to predict treatment responses. Customized chemotherapies based on certain biomarkers have been shown to have better efficacy and result in improved outcome in cancer patients. Ab initio determination of tumor biomarkers may allow for a more sophisticated strategy for treatment design, and hopefully, a more effective therapeutic response. Pharmacogenetics analysis of tumor tissues is another field that may provide promising prognostic and/or predictive values, especially in identifying important gene signatures responsible for risk assessment, tumor response, and subsequent clinical outcomes.
Previously, we have had preliminary information regarding certain biomarkers in pancreatic cancer such as cytidine deaminase’s role in degrading gemcitabine efficacy, several EGFR pathway markers as potential targets in erlotinib therapy, as well as germ line mutations leading to various single nucleotide polymorphisms (SNPs) causing different tumor biological responses to standard chemotherapy. However, detailed information from large or randomized phase III trials is yet to be revealed.
In this review, we will provide updates on some of these biomarkers: we will show certain SNPs with concordant interaction with gemcitabine-based therapy identified through a large study using genome-wide interaction analysis; we will introduce the concept that circulating tumor cells can be a useful prognostic factors in progression free survival; we will also present new confirmatory findings of cytidine deaminase’s role in gemcitabine-based chemotherapy, as well as several EGFR pathway markers analysis in pancreatic cancer treated with erlotinib.
Table 1 summarizes the related abstracts and their main findings.
Genome-Wide Single Nucleotide Polymorphisms (SNPs) Identification for Concordant Interaction with Gemcitabine Chemotherapy
The traditional chemotherapy for pancreatic cancer is primarily gemcitabine-based. However, recent study has recommended other regimens such as oxaliplatin and irinotecan plus fluorouracil and leucovorin (FOLFIRINOX) [1], which confers superior efficacy but carries higher toxicities [2]. As such, it is desirable to identify those patients who will benefit the most from traditional gemcitabine-based therapy to minimize unnecessary toxicities-associated morbidities. McWilliams et al. (Abstract #4022 [3]) was able to present a study identifying 9 SNPs as good predictive markers for gemcitabine therapy, by using a genome-wide interaction study on a large cohort of 940 patients. In their study, initial 550,000 SNPs from 413 patients of all stages of pancreatic cancer treated with gemcitabine were screened for treatment-benefit interaction with overall survival being the endpoint. The 100 most significant SNPs from initial stage were further analyzed on another 527 patients for concordant effects from gemcitabine therapy. The authors were able to identify a total of nine such SNPs that have statistically significant favorable predictive power in gemcitabine efficacy. Specifically, genes PYCARD (rs6507115) and MAPRE2 (rs8056505) were strongly correlated with gemcitabine efficacy in both stages, which makes them potential genomic predictive biomarkers. No SNPs has been found to have significant association with gemcitabine toxicities.
Circulating Tumor Cells (CTCs)
Circulating tumor cells (CTCs) are cancer cells that are detached from primary tumor sites and travel in the peripheral blood circulation system, leading to distant metastasis [4] (Figure 1). CTCs are typically enriched and detected via immunomagnetic separation system [5] or via microfluidic CTC-chip system [6, 7]. Both adenocarcinoma [6] and neuroendocrine [8] pancreatic CTCs have been detected. Khan et al., had shown original preliminary results suggesting the prognostic and predictive values of CTCs in neuroendocrine pancreatic cancer [8].
De Albuquerque et al. (Abstract #e14657 [9]) reported the prognostic values of CTCs detection in pancreatic adenocarcinoma. By using the high affinity antibodies BM7 (MUC 1) in addition to conventional VU1D9 (EpCAM), CTCs detection were reported in 49.3% of 144 peripheral blood samples from 39 patients with advanced pancreatic adenocarcinoma. The detection of such CTCs portended poor prognosis (median progression free survival: 60.7 days vs. 163.6 days in patients with positive CTCs detection and negative CTC detections, respectively; P<0.0001). As such, authors concluded that CTCs can act as an independent prognostic biomarker.
Cytidine Deaminase (CDA) and CDA*3
Cytidine deaminase (CDA) genotype and phenotype as negative predictive markers for gemcitabine was further elucidated. Gemcitabine is deactivated by CDA to an inactive form, 2’,2’-difluorodeoxyuridine (dFdU), during its biotransformation [10] (Figure 2). CDArelated biomarkers predicting gemcitabine-induced toxicities, as well as negative efficacy, have been previously reported [11]. Conversely, it has also been reported that homozygous CDA*3, a non-synonymous SNPs of CDA (208 G>A) is associated with very low plasma CDA level and, thus, largely induces high level of gemcitabine and its associated toxicities [12].
Figure 2. Gemcitabine biotransformation (adapted from Gilbert et al. [10]).
DCK: deoxycytidine kinase; dFdC: gemcitabine; dFdCDP:
gemcitabine diphosphate; dFdCMP: gemcitabine monophosphate;
dFdCTP: gemcitabine triphosphate; dFdU: 2′,2′-difluorodeoxyuridine;
dFdUMP: 2′,2′-difluorodeoxyuridine monophosphate; UMPCMPK:
deoxycytidylate kinase.
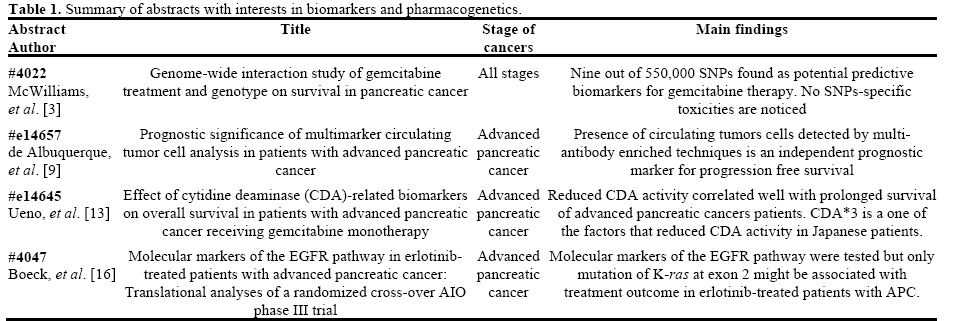
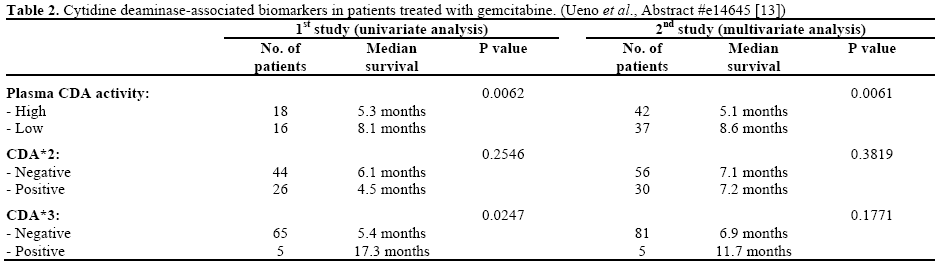
Ueno et al. (Abstract #e14645) reported their results of CDA-related phenotypic and genetic biomarkers on the overall survival of advanced pancreatic cancer patients treated with gemcitabine monotherapy [13]. Markers including mean residence time of gemcitabine, plasma concentration of dFdU at 0.5 h (C0.5), plasma CDA activity, as well as CDA*2 and CDA*3 genotype, were studied. Results from two studies were reported.
During their first study (n=73), univariate analysis was applied between biomarkers and overall survival. Significant survival advantage was found in patients who have longer mean residence time and higher C0.5 (P=0.0138 and P=0.0011, respectively). Accordingly, low plasma CDA activity and heterozygous CDA*3 were also significantly associated with prolonged overall survival (P=0.0062 and P=0.0247, respectively). In a second study (n=98), where multivariate Cox proportional hazard model was used, low plasma CDA level was again confirmed to be associated with longer overall survival and positive trend between CDA*3 heterozygosity and overall survival was observed though statistical significance was not achieved (Table 2). No statistically significant associations between CDA*2 and survival benefits were observed in either studies.
Molecular Markers of the EGFR Pathway in Erlotinib- Treated Patients with Advanced Pancreatic Cancer
Gemcitabine plus erlotinib, EGFR tyrosine kinase inhibitor, has been approved by U.S. Food and Drug Administration as a first line therapy for patients with locally advanced or metastatic pancreatic cancer as statistically significant overall survival and progresssion- free survival have been reported in a phase III double-blind trials (n=569) [14]. To further assess if potential molecular biomarkers in the EGFR pathway can be identified, retrospective analysis was performed on the Arbeitsgemeinschaft Internistische Onkologie (AIO) phase III trial [15] where similar overall survival was ascertained between patients with advanced pancreatic cancer receiving either capecitabine plus erlotinib followed by gemcitabine monotherapy or gemcitabine plus erlotinib followed by capecitabine monotherapy.
The authors (Abstract #4047 [16]) performed tissue analysis on the archived formalin fixed paraffin embedded tumor tissues on 208 patients. The followings markers were included in the analysis: Kras exon 2 mutation status, EGFR expression, PTEN expression, EGFR intron 1 polymorphism, and EGFR exon 13 R497K polymorphism. About 70% of patients are found to have K-ras mutations while 96% showed EGFR expression by immunohistochemistry. In univariate biomarker analyses, only K-ras mutations status was found to have significant associations with overall survival benefit (hazard ratio: 1.68; 95% CI: 1.17-2.41; P=0.005). Additionally, none of the aforementioned biomarkers were found to have statistically significant association with skin rashes, a common side effect from erlotinib. Therefore, definitive translation data is still relatively limited from this study, though authors have proven the feasibility to perform retrospective biomarker analysis on archived tissues data from this AIO phase III trial.
Genome-wide analysis using high-throughput DNA method for potential molecular biomarker identifications and analysis is an attractive strategy in pharmacogenetics. Given the lethality and complexity of pancreatic cancer, reliable pharmacogenetic profiling is yet to be determined. In this review, we present a promising set of SNPs, such as PYCARD and MACRE2, which appears to have strong positive correlation with efficacy from gemcitabine-based chemotherapy in pancreatic cancer. These preliminary data may provide ground work and rationale in designing future clinical trials, further elucidating and possibly validating such biomarkers. The challenge and the dilemma we always face when new technologies are involved is regarding cost, accessibility and all the other potential laboratory problems might arise, from reliability to staff availability, etc..
We also introduced the concept of using circulating tumor cells as prognostic biomarker in pancreatic cancers. The prognostic and predictive values of circulating tumor cells have been well established in breast and prostate cancer, though their utility in pancreatic cancer is very limited. As far as we know, the abstract we reviewed presented the first set of data supporting the potential role as a biomarker of CTCs in pancreatic adenocarcinoma. We tend to agree with original authors that, as technologies further advance, it is possible that CTCs may emerge as a critical prognostic as well as predictive biomarkers in pancreatic cancer, but there is still a long way to go.
The predictive value of cytidine deaminase and its associated phenotype and genotype in determining the efficacy of gemcitabine is re-visited. As the results from the two small studies presented this year were not similarly conclusive, prospective validation in larger trials are needed. Similarly, identification of a predictive biomarker for the EGFR inhibitor erlotinib on retrospective tissue analysis of the previous AIO phase III trial failed, though a mutation of K-ras at exon 2 may play a role, but further research is definitely warranted.
Therefore, in view of data available, our clinical practice remains unchanged, though some of aforementioned biomarkers appear to have a potential prognostic and predictive role and have to be explored further. Given these promising preliminary data, future clinical trials using hybrid chemotherapy design [17], tailored towards standardized biomarker assay, may bring forward more insight and confirmatory data for this interesting concept.
The authors have no potential conflicts of interest