Original Article - (2018) Volume 19, Issue 3
1Department of Radiation Oncology, Beaumont Health, Royal Oak, Michigan, USA
2Beaumont BioBank, Beaumont Health, Royal Oak, Michigan, USA
3Department of Human Oncology, Carbone Cancer Center, University of Wisconsin, Madison, WI, USA
4Department of Pathology, Beaumont Health, Royal Oak, Michigan, USA
5Department of General Surgery, Beaumont Health, Royal Oak, Michigan, USA
Received March 19th, 2018 - Accepted May 09th, 2018
Objectives The ability to identify patients that have disease progression soon after surgical resection could guide treatment as well as aid in the development of novel targeted therapies. This study correlates gene expression and overall survival in patients with pancreatic adenocarcinoma. Methods Patients with pancreatic adenocarcinoma treated with definitive surgery without neoadjuvant therapy were grouped into short-term (<10 months, n=13) and long-term (>20 months, n=11) survivors. Ribonucleic acid was extracted from snapfrozen tissues, and global gene expression was examined. Pathway analysis was also performed. Results The mean overall survival in each group was 7.5 and 32.0 months. We identified 163 genes that were differentially expressed between patients who survived <10 months and >20 months after definitive surgery. Many of the genes identified have known prognostic importance; however, less than half of these genes have been reported to be associated with survival in pancreatic adenocarcinoma. Pathway analysis identified expression targets of SP1, JUN, and EGF to be highly regulated based upon differences in overall survival. Conclusions In pancreatic adenocarcinoma patients who have undergone definitive resection, we have identified multiple genes associated with inferior survival. Many of the genes reported in this study have not previously been linked to overall survival in this patient population.
Gene Expression; Pancreatic Neoplasms; Survival
CSCs cancer stems cells; PDAC pancreatic ductal adenocarcinoma; RNA Ribonucleic acid
Pancreatic cancer is a challenging disease with dismal prognosis for the vast majority of afflicted patients. The incidence of pancreatic cancer in the United States is estimated to be 48,960 in 2015 with 40,560 deaths and a 5-year survival rate of 7% [1]. Pancreatic ductal adenocarcinoma (PDAC) accounts for about 90% of the malignant cancers arising from the ductal epithelium in the exocrine part of the pancreas gland. Due to the silent nature of the disease, most patients present late with unresectable disease but approximately 20% will undergo resection followed by chemotherapy and radiation treatment. Disappointingly, despite the use of adjuvant therapy, a significant number of these patients will recur early after resection and die of the disease within one year [2] whereas 25% of these patients with resected PDAC can live for 5 years or more [3]. Traditional prognostic factors including stage, tumor grade, negative surgical margins, and absence of lymph nodes cannot always accurately predict long-term survival. The biology of the tumor may be more important in predicting distant recurrence and ultimately survival. Identifying prognostic factors that can predict which patients may live longer would help with treatment decisions postoperatively but also including the use of neoadjuvant therapy as a means of delaying surgery in patients who would otherwise have rapid disease progression.
One way to select these individuals would be to define a prognostic signature that can identify patients with more aggressive tumor biology prior to treatment. Many different aspects of PDAC tumor biology have been suggested as candidates for determining the aggressive phenotype including genetic [4], epigenetic [5], tumor microenvironment [6], immune response [7] or presence of cancer stem cells (CSCs) [8]. A recent report failed to find a difference in the somatic mutation profile of PDACs in very long-term survivors compared to PDACs in patients unselected for survival [9]. In the absence of a specific genetic mutation profile that discriminates long-term survivors, another approach is to study gene expression. Expression profiling of PDAC has been undertaken by several different investigators [10, 11] and uncovered various signaling pathways associated with tumor progression and metastatic disease. Of particular interest is the study of Stratford et al. [12] who discovered a sixgene signature that was predictive of survival in localized PDAC in comparison to metastatic PDAC. Interestingly, most genes in the classifier (SIGLEC11, KLF6, NFKBIZ, ATP4A, GSG1, and FOSB) did not have an obvious role in carcinogenesis, and only three had significantly higher expression in the poor prognostic patients.
Instead of comparing primary PDAC tumors at the extremes of disease (localized versus metastatic), we specifically selected a subgroup of patients who were all considered candidates for surgical resection, and from this cohort we further selected patients with short-term (<10 months) and those with long-term survival >20 months). Our focus was not only to identify genes of interest so as to assist in the development of a specific prognostic gene signature that could guide treatment decisions at presentation and postoperatively but also to identify the key pathways involved in patients with poor outcomes as potential targets for novel treatment strategies.
Patient Consent and Sample Acquisition
Between February 2009 and November 2013, patients who underwent a pancreaticoduodenectomy for pancreatic adenocarcinoma were approached to submit portions of their tumor to the Beaumont BioBank. A single surgeon completed all resections. In addition to surgery, patients received adjuvant therapy as previously reported [13]. Patients were consented by Beaumont BioBank clinical staff using an IRB approved protocol (HIC 2008-180), and samples were processed and stored at -80°C using standard operating procedures. Inclusion criteria required survival of greater than 100 days following surgery in order to eliminate death due to surgical complications or other comorbidities. Analysis was limited to patients who did not present with distant metastasis and did not receive preoperative chemo- or radiation therapy. Resection margins were assessed using a standardized pathology protocol based on axial specimen slicing and reporting margin involvement if tumor cells are present within 1-2 mm from the margin. 39 patients were consented and their specimens banked and of these 32 satisfied the inclusion criteria. The overall median survival of all patients was 10.1 months. For this study, we were specifically interested in whether there were genomic differences between the longer-term survivors and those with shorter survival. Therefore, we arbitrarily selected two sub-groups of patients. We identified 11 patients who lived greater than 20 months following their surgery and 13 patients who lived less than 10 months. This lower cut-off was chosen based on the median survival and the upper cut-off of 20 months was simply a doubling of the median survival and also gave a similar number of patients in each group.
RNA Isolation
Frozen pancreatic adenocarcinoma tissue specimens stored at -80°C in RNAlater Stabilization Solution (Life Technologies, Carlsbad, CA) were homogenized into lysis buffer using the gentleMACS dissociator’s (Miltenyi Biotec Inc., Auburn, CA) “Homogenization of tissue for total RNA isolation” protocol. Following the manufacturer’s protocol, RNA was purified using the E.Z.N.A. Total RNA Kit I (Omega, Norcross, GA), quantified (Nanodrop 8000, Thermo Scientific), and then stored at ‑80°C. RNA integrity was determined by Bioanalyzer analysis (Agilent) just prior to processing for expression microarray analysis.
Illumina Expression Beadchips
RNA was amplified and labeled using the TargetAmp- Nano Labeling Kit (Epicenter, Madison, WI) which enables amplification and target preparation compatible with the Direct Hybridization Assay (Illumina, San Diego, CA). Amplification was performed with 500 ng of total RNA input following procedures described in the TargetAmp- Nano Labeling Kit user guide. Hybridization and staining to the HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA) was performed using 750 ng of BiotinaRNA product following protocols outlined in the Whole- Genome Gene Expression Direct Hybridization Assay Guide. Subsequent scanning of the BeadChip was performed using the iScan Microarray Scanner (Illumina, San Diego, CA).
Gene Expression and Pathway Analysis
Gene expression data from 24 samples were imported into Illumina’s Genome Studio. They were imported with cubic spline normalization. Quality control was performed in Genome Studio. The Partek Report Plug-in from Illumina’s Genome Studio was used to export the gene expression data from 26 arrays into Partek’s Genomics Suite (version 6.15.1207). Differentially expressed genes were detected by ANOVA (p≤0.01 and 2-fold cutoff) taking into account the parameters of survival and barcode. Barcode refers to the chip used; it is included to account for hybridization differences associated with runs on different bead chips. Pathway analysis was done with Pathway Studio (Elsevier, version 11.1.0.6 2015-12-08). The data are available using NCBI Gene Expression Omnibus accession number GSE77435 [14].
Survival Analysis
Selected genes were analyzed for their influence on survival from pancreaticoduodenectomy. The log 2 intensity values for the long and short-term survivors were used to classify patients as above and below the median value and analyzed using the Kaplan-Meier method and log rank test. A p-value of <0.05 was considered statistically significant. Statistical analyses were performed SPSS (version 22; IBM SPSS, Amok, NY, USA)
Patient Clinical Data
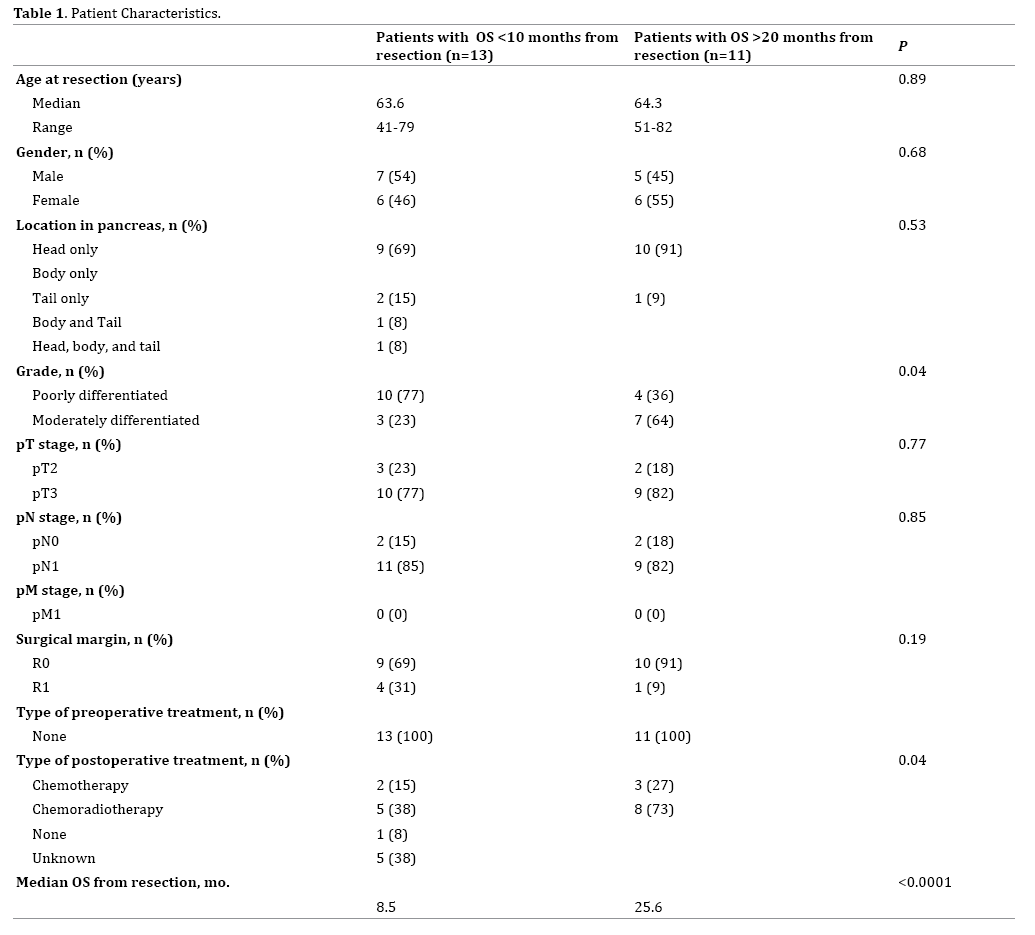
Table 1 lists patient characteristics, pathologic findings, additional treatments, and overall survival (OS) from the time of surgery. The median age was 64 years, and 50% of patients were male. The median OS after surgery for all patients in the study was 10.1 months with a median of 25.6 months and 8.5 months for the >20 months and <10 months groups, respectively. There was no significant difference in the pathologic T (T3=77% vs. 82%; p=0.77) or N (N1=85% vs. 82%; p=0.85) stage or positive surgical margins (31% vs. 9%; p=0.19) between patients based upon OS. However, there was significantly higher proportion of grade 3 tumors in the short-term survivors compared to the long-term survivors (77% vs. 36%; p=0.04). There was also a difference in postoperative (p=0.04) treatment.
Gene Expression Differences At >20 Months
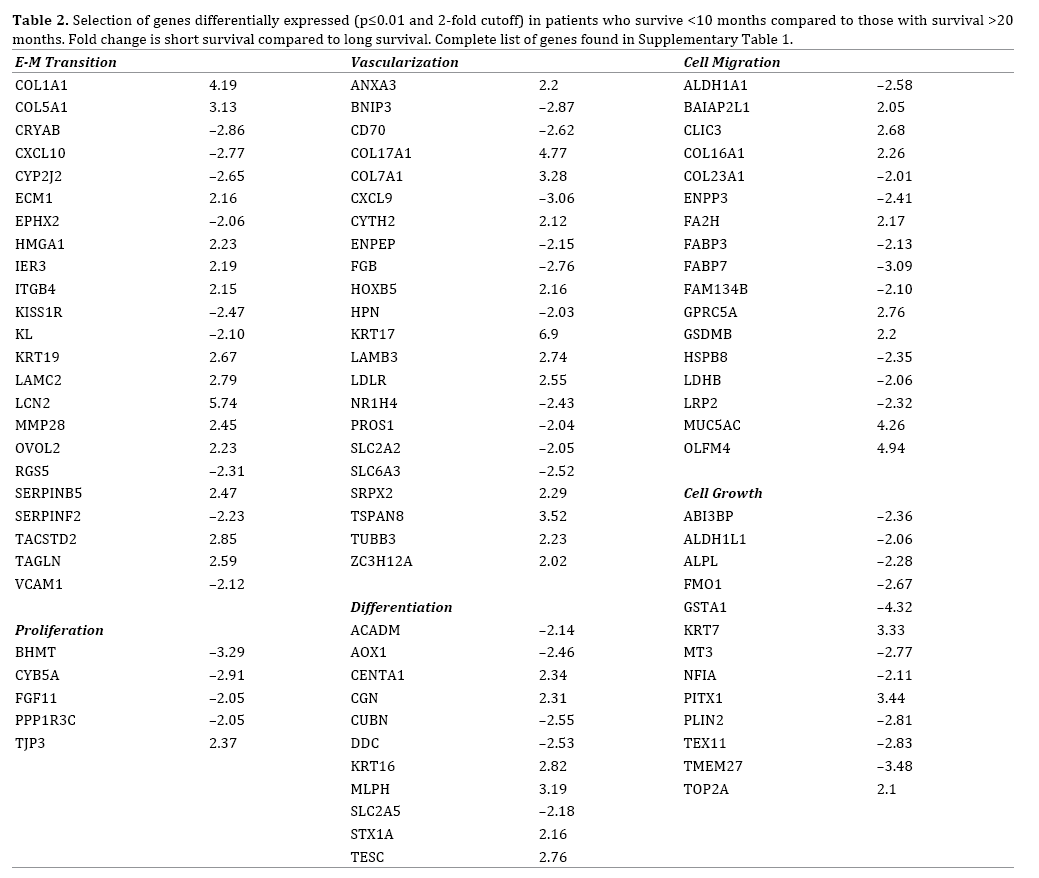
We identified 163 genes that were differentially expressed (p≤0.01 and 2-fold cutoff) between patients who survived <10 months and patients with survival >20 months (Supplemental Table 1). This included genes associated with epithelial to mesenchymal transition, vascularization, and cell migration (Table 2). Some of the greatest increases in expression for short-term survivors were seen in KRT17, S100P, LCN2, COL17A1, and COL1A1 amongst others whilst some of the most prominent genes that were downregulated in the long-term survivors included GSTA1, GSTA2, LGALS2, and CXCL9.
Signaling Changes At >20 Months
Pathway Studio utilizes a literature mining tool, MedScan, to generate sub-networks that associate genes with other entities such as cell processes. A Fisher’s Exact test was used to identify sub-networks that are highly represented by differentially expressed genes. One type of sub-network associates genes with a central seed based upon the ability of the seed to control gene expression. The top expression target sub-networks that were highly represented by the differentially expressed genes included the expression targets of SP1, JUN, and PPARG (Figure 1).
Figure 1. Combined expression target sub-networks of SP1, JUN, and PPARG identified as highly represented in the list of genes differentially expressed (p≤0.01 and 2-fold) between patients who survived <10 months and those that survived >20 months. Genes in red are upregulated in patients with shorter survival; genes in blue, downregulated.
Of note is the general downregulation of PPARG expression targets and the presence in these sub-networks of COL1A1, COL7A1, GPRC5A, KRT17, and ECM1 which have not been previously linked to patient outcomes in PDAC. Another type of sub-network includes genes involved in regulating cell processes. Differentially expressed genes between patients with OS <10 months and those with OS >20 months are highly represented by genes regulating cell differentiation, cell proliferation, cell migration, and vascularization.
Survival Analysis
Figure 2a shows the Kaplan-Meier survival curves for ALDH1A1, COL17A1, COL7A1 and KRT17 selected from Table 1 and 2. COL17A1 and COL7A1 were the most significant genes in terms of overexpression being linked to poor survival (Figure 2b, 2c) whilst ALDH1A1 was the most significant gene where underexpression was linked to shorter survival (Figure 2a).
Figure 2. Kaplan-Meier survival analyses of selected genes. The green solid lines represent tumors with lower than median expression whilst the red dotted lines are those with higher than median expression. The genes shown are (a). ALDH1A1, (b). COL17A1, (c). COL7A1 and (d). KRT17. The p-values represent the Log-Rank test.
This study reports on pancreatic adenocarcinoma gene expression differences in patients who survived greater than 20 months or less than 10 months following surgery. There was no significant difference in the age, sex, or stage between the two groups, and no one received preoperative therapy. There was, however, a significantly greater percentage of high grade tumors in patients who lived <10 months.
Traditional prognostic criteria for long-term survivors have included negative margin status, small tumor size, no lymph node involvement, low CA 19-9 level, low grade, absence of metastases, and type of treatment administered [15, 16, 17]. However, the use of these prognostic factors has limited value due to the heterogeneity of the long-term survival group. Ferrone et al. showed that negative margins and negative nodes demonstrated a positive prognosis, but at the same time 41% of long-term survivors had positive nodes and 24% had positive margins [18]. Adham et al. found that typically positive prognostic criteria did not predict long survival with 29 of 30 long-term survivors having T3/T4 tumors with 12 of 30 having positive lymph nodes [19].
One possible conclusion is that the biology of the tumor, not traditional prognostic markers, is important for prediction of long-term survival. One recent study by Dal Malin et al. tried to address this by using next-generation exome sequencing to examine the genomic profile of longterm survivors [9]. While mutations were found in KRAS, TP53, SMAD4, and CDKN2A, there were no mutations that were preferentially found in the long-term survivors.
In order to continue the search for the biological variability seen in the long-term survivors, we have identified 163 genes that were differentially expressed between the <10 months and >20 months survival groups. Several of the genes we identified have a known prognostic role in pancreatic adenocarcinoma including ADAM metallopeptidase domain 8 (ADAM8) and transgelin (TAGLN) along with aldehyde dehydrongenase 1 family, member A1 (ALDH1A1). However, most of the genes we identified have not been previously linked to prognosis in pancreatic adenocarcinoma.
Aldehyde dehydrogenase 1 family, member A1 is an enzyme involved in alcohol metabolism. It was found to have decreased expression (2.6-fold) in short-term survivors and was significant in overall survival analysis. This gene has been previously linked to prognosis and progression in many cancers [20, 21, 22, 23]. Expression of this gene has also been linked to cancer stem cells (CSCs), with low expression associated with gemcitabine resistance and poor prognosis in pancreatic adenocarcinoma [24, 25, 26].
Keratin 17 is an intermediate type I filament chain keratin usually expressed in the nail bed, hair follicle, and sebaceous glands. This gene was found to have higher expression (6.9 fold) in in the short-term OS patients and was significant in overall survival analysis. Increased expression of this gene has been linked to poor survival in cervical squamous cell carcinoma, epithelial ovarian cancer, gastric cancer, and breast cancer [27, 28, 29, 30]. However, this has not been previously demonstrated to be prognostic in pancreatic cancer.
Several members of the COL family were associated with short survival. The COL (collagen) family comprises 28 members that contain at least one triple-helical domain. Collagens are deposited in the extracellular matrix (ECM) where most of them form supramolecular assemblies. They are known to affects the tumor microenvironment through degradation and re-deposition contributing to ECM remodeling which promotes tumor infiltration, angiogenesis, invasion and migration. While collagen was traditionally regarded as a passive barrier to resist tumor cells, it is now evident that collagen is also actively involved in promoting tumor progression [31]. COL17A1overexpression was the most significant gene in terms of overexpression being associated with shorter survival. This gene has also been recently identified as a potential biomarker using a minimum-redundancymaximum- relevance (mRMR) method interrogating a set of transcriptome data of pancreatic cancer [32].
This study adds to the growing literature on the prognostic utility of gene expression patterns for PDAC. The University of Virginia recently published a 13-gene signature that predicts significantly higher risks of mortality in pancreatic adenocarcinoma patients [33]. Of the 13 genes these authors identified, 4 including TGFA, ELAVL1, and MDM2 had been previously shown to be important in pancreatic adenocarcinoma, 6 genes were associated with prognosis or highly expressed in other forms of cancer but not previously reported in PDAC, and 3 had not been reported to be prognostically significant in any malignancy. Those categorized as low risk by this gene signature had a median overall survival of 14 months compared to 6 months for high risk patients. There was no overlap in the genes identified for their gene signature and the genes identified in the current study. A 6-gene prognostic signature was also published by the University of North Carolina [12]. This signature included FOSB, NFKBIZ, IKBZ, MAIL, GSGI, and SIGLEC11. Patients classified as low risk by this study had a median overall survival of 49 months, and those classified as high risk had a median overall survival of 15 months.
Additionally the current results were compared to a pair of studies in NCBI’s Gene Expression Omnibus. The original purpose of these published studies was not to examine survival specifically, but both studies included survival data for at least some of the publicly available data. In a study by Van den Broeck et al. [34], microarrays (GEO accession GSE42952) were used to compare patients with good (DFS>50 months) and poor (DFS<7 months) outcome. From a study by Yang et al. [35], the data from a subset of patients (GEO accession GSE62452) were used to compare patients with similar outcomes as the current study – patients with OS>20 months or <10 months. Of the genes that were identified as differentially expressed in the current study (Supplemental Table 1), 56 were validated through the analysis of the data in these studies. This included genes such as COL1A1, COL5A1, COL7A1, CYB5A, and STX1A that were confirmed by both studies. Even more striking was the concordance in regulated cell processes between our current study and the publicly available data. Of the highly regulated cell processes that we identified, five of the top six were found in both external studies. This included cell differentiation, cell invasion, cell proliferation, cell migration, and cell behavior which were all highly ranked in both publicly available datasets.
In addition to individual gene expression biomarkers, signaling surrounding SP1, JUN, and EGF was highly altered in the patients that showed longer survival. This result confirmed the role of these signaling pathways in pancreatic cancer. SP1 is a negative prognostic factor that plays a role in cell proliferation and metastasis [36]. In particular, SP1 protein was found to be overexpressed in a subset of primary pancreatic adenocarcinoma that developed lymph node metastasis, were higher stage and grade, and had a much shorter overall survival [37]. JUN is a transcription factor involved with cell proliferation that has been identified as an oncogene. It has previously been related to pancreatic cancer stage, grading, and invasion [38]. Expression of JUN was shown to be elevated in liver metastases compared to pancreatic cancer tissue, and high expression was seen more often in short-term survivors [39]. EGF acts via its receptor (EGFR) to potentiate growth, proliferation and differentiation of many different cell types. Specifically, it has been shown to be involved in growth, invasiveness, and metastasis of pancreatic cancer [40, 41].
In this comparison of patients who live <10 months and >20 months following definitive resection for pancreatic adenocarcinoma, we have identified multiple differentially expressed genes. Some of these genes have previously been shown to be prognostic in pancreatic adenocarcinoma, but most had never been linked to survival in these patients. These genes and their expression targets warrant further investigation to determine their value as prognostic markers or targets for molecular therapy.
This work was funded by the Mopper Family philanthropy fund.
The authors declare that they have no conflicts of interest.