Keywords
Immunogenic; CRISPR/Cas9 system; Homology-directed repair
Introduction
CRISPR/Cas9 genome-editing system comprises an enzymatic machinery or "molecular scissors” Cas9 that causes site-specific DNA double stranded breaks (DSB) in a sequencespecific manner using a guide sequence within an easy-tosynthesize tracrRNA–crRNA duplex molecule [1-12] (Figure 1).
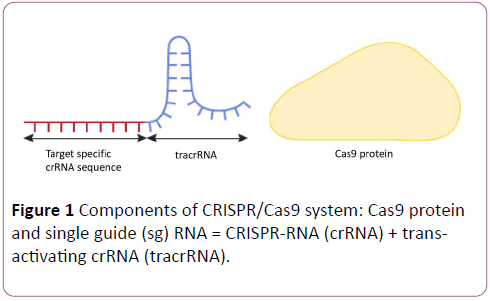
Figure 1: Components of CRISPR/Cas9 system: Cas9 protein and single guide (sg) RNA = CRISPR-RNA (crRNA) + transactivating crRNA (tracrRNA).
CRISPR/Cas9 is mainly used to either disrupt genes or correct specific genes by inserting specific DNA sequences to replace the defective ones. In both approaches, the cell repair machinery rejoins the ends via either non-homologous end joining (NHEJ) or homology-directed repair (HDR) (Figure 2). CRISPR/Cas9-mediated knockout is usually preferred rather than inserting or replacing sequences because the latter requires a very adequate HDR pathway [13-16].
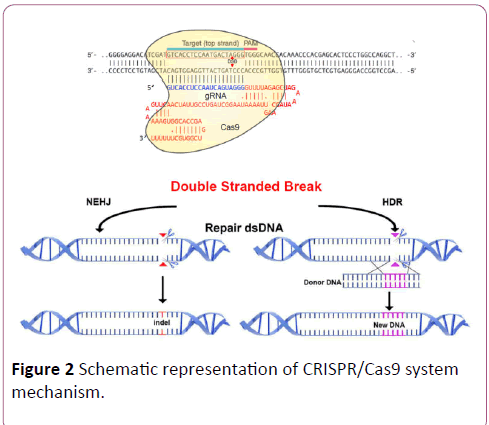
Figure 2: Schematic representation of CRISPR/Cas9 system mechanism.
CRISPR/Cas9-mediated gene therapy components could be delivered in vivo or in vitro. in vivo, it can be delivered directly into tissues by using the most common adeno-associated virus (AAV) vector procedure [17-20], or non-viral-based pathways such as hydrodynamic injection [21,22].
The non-viral-based delivery of CRISPR/Cas9 via hydrodynamic injection was employed in to correct tyrosinaemia-causing mutation, but it was ineffective in humans [23]. The ex-vivo procedure is usually performed by either the electroporation-mediated transfection technique, which enables CRISPR components to be easily delivered to difficult-to-transfect cells such as human T-cells [24]. The electroporation-mediated transfection technique was successfully used to restore dystrophin expression of skeletal muscles in Duchenne muscular dystrophy (DMD) in mice [25]. The ex-vivo delivery can also be performed by obtaining adult stem cells or fibroblasts from patients to be cultured and reprogrammed, and then to be corrected by CRISPR/Cas9 and eventually reintroduced to the patient. This approach circumvents some difficulties in delivering these novel geneediting therapies to the right tissues, such as delivering CRISPR/Cas9 to target β-globin gene for treatment of β- thalassaemia [26].
Recent Advances and Applications
CRISPR/Cas9 has helped to programme transcriptional factors to control gene expression by performing homozygous knock-in or biallelic knockout in order to create transgenic or disease models such as in gain- or loss-of-function studies [27,28]. We can also alter gene expression by using a dead or broken Cas9 enzyme to block the binding of RNA polymerase needed for the gene to be expressed or attaching a dead or broken Cas9 protein to transcriptional factors or activator to stimulate the gene expression [29,30]. Furthermore, CRISPRCas9 was used to construct somatic and germline mouse models with point mutations or chromosomal deletions in multiple genes using multiple gRNAs, and even more complex chromosomal rearrangements. Researchers successfully integrated large transgenes via AAV vectors into primary human T cells, CD34+ hematopoietic stem and progenitor cells [31,32]. Such models can also be used to broaden our understanding of the progression of specific tumours and help identify new approaches for cancer therapy [33]. Unlike conventional RNA interference, which cleaves only dsRNA using Dicer enzyme with high off-target effects and low reproducibility [34], reversible CRISPR/Cas9-mediated transcriptional repression can target any specific region throughout the whole genome with more predictable offtarget effect [35-38]. In the field of epigenetics, a dead Cas9 could be attached to epigenetic modifiers to activate DNA methylation by adding methyl group to DNA or to modify histones by adding acetyl groups to histone proteins, making epigenome reprogrammable and furthering our understanding of how a specific modification influences gene expression [30].
Scientists successfully modulated human CD4+ T-cells in vitro by disabling a protein that is used by HIV to infect these cells [24]. They were also able to insert a specific DNA sequence to modify the expression of PD-1 that enables T-cells to attack tumours. This has potential therapeutic applications not only in the treatment of HIV, but also cancer and autoimmune disorders. However, delivering cas9 and singleguide RNA via an AAV vector was insufficient to modify protein expression in T-cells in vivo. An effective solution was found by editing the T-cells in vitro and these cells were then returned to the patient [24]. Similarly, by applying a lentivirus-delivered RNA-guided CRISPR/Cas9 system ex-vivo on cultured infected CD4+ T-cells from patients with AIDS at the latent stage, researchers successfully halted viral replication by excising the whole host-integrated HIV-1 genome copies from the infected CD4+T-cells [39]. Extensive examination using diverse methodologies showed sustained therapeutic effect and decreased toxicity compared with DNA-based delivery and there were no off-target effects or complications regarding various biological functions of patients’ cells [39]. They thereby established an alternative evolutionary therapy to expensive lifetime antiretroviral treatment. However, individualized CRISPR/Cas9 systems are needed due to the supposed versatility of different integrated HIV-1 sequences with the CD4+ T-cells of different patients [40].
A breakthrough was made by three separate groups who effectively corrected Duchene muscular dystrophy in mice by excising one or more exons from the DMD gene to restore the reading frame disrupted by frameshift deletion or duplication mutations in the DMD gene [13,41-43]. Snipping out exon23 resulted in shortened yet functional dystrophin protein in muscles of various organs, which reduced disease severity, and more importantly in the myocardium, which lessened the risk of premature fatal heart failure [13]. In one study, systemic delivery of CRISPR/Cas9 helped dystrophin expression increase over time due to positive selection for the edited cells phenotype, which means that the effect is maintained for a long time [43]. Exhaustive evaluation of the functions of the whole genome was performed to test the specificity and efficacy of this system by robust methodologies like PCR and Sanger sequencing which confirmed the induced excision. Moreover, different biochemical, immunoassay, histological and morphological examinations showed that dystrophin expression was partially restored and the muscular function improved in most vital organs [43]. This provides convincing evidence about the efficacy of one-time administration of AAVCRISPR- mediated therapy in Duchene muscular dystrophy in new-born and adult mice. Inexplicably, the systemic delivery of CRISPR/Cas9 in one study resulted in no observed improvement in the muscle function of distal extremities [13]. Even though there were no observable off-target effects in animal models, the small number of in-silico predicted sites does not cover the potential off-target activities in vivo in humans. Therefore, the in vivo safe delivery of this therapy should be evaluated using other extensive measures, such as wide-genome assessment, before such experiments can be developed into therapeutics in the clinic [41]. More interestingly, in Duchenne muscular dystrophy caused by exons duplication, snipping out the duplication of DMD exons 18–30 restored the full-length of dystrophin [44].
Furthermore, researchers have successfully restored the normal activity and functionality of cystic fibrosis transmembrane conductor receptors (CFTR) in human intestinal organoids by correcting mutations in the CFTR locus in cultured intestinal stem cells using CRISPR/Cas9-mediated homologous recombination followed by delivering the product to the patient’s colon [45-47]. However, as cystic fibrosis can affect multiple organs, this approach has clear limits. In an experiment to correct mutations in Fah gene in hepatocytes of a mouse model for the treatment of tyrosinaemia and in order to deliver CRISPR system into the liver to correct the mutant gene by only 0.04%, they needed to pour large quantities into the circulation, which is not feasible [23].
CRISPR/Cas9 has also been efficiently used to correct β- thalassaemia via ex-vivo editing of mutations in the patientspecific primary somatic stem cells or iPSCs and these edited cells could be transplanted in the patient’s bone marrow [48]. These findings encouraged researchers under the existing rules in China to cross the ethical boundaries and try to correct mutations in β-globin (HBB) gene in human tripronuclear zygotes. However, mosaicism and off-target cleavages occurred [49]. The main advances and applications of CRISPR/Cas gene editing were summarized in Table 1.
| Applications |
Refernces |
| Repair of CFTR in intestinal stem cell organoids of patients with cystic fibrosis |
Schwank, Gerald, et al. (2013) |
| Correction of a Fah mutation in hepatocytes in a mouse model for the treatment of tyrosinemia |
Yin, Hao, et al. (2014) |
knockout for genes involved in resistance
to the BRAF protein kinase inhibitors in human cells for the treatment of melanoma |
Shalem, Ophir, et al. (2014) |
| Gene correction of ß-thalassemia mutations in patient-specific iPSCs |
Xie, Fei, et al. (2014) |
| Correcting Crygc gene in cataracts in mouse spermatogonial stem cells |
Wu, Yuxuan, et al. (2015) |
| "knock in" targeted genome modifications to modulate T-cell primary human CD4+ T-cells in treatment of HIV |
Schumann, Kathrin, et al. (2015) |
| Gene editing of ß-thalassemia mutations in human tripronuclear zygotes |
Liang, Puping, et al. (2015) |
| CRISPR/Cas9 ß-globin gene targeting in human haematopoietic stem cells in treatment of sickle cell disease and ß-thalassaemia |
Dever, Daniel P, et al. (2016) |
| Integration of a super-exon into the CFTR of cystic fibrosis cell lines |
Bednarski, Christien, et al. (2016) |
| Elimination of HIV-1 genomes from human CD4+ T-cells |
Kaminski, Rafal, et al. (2016) |
| Genome editing in chronic Hepatitis B virus (HBV) infection |
Seeger, Christoph, and Ji A. Sohn. (2016) |
| Gene correction of cardiomyopathy in mice |
Carroll, Kelli J, et al. (2016) |
| Correcting Alzheimer’s Disease-associated mutations in APOE4 in mouse astrocytes and TP53 mutations in a mouse breast cancer line |
Komor, Alexis C, et al. (2016) |
| Correction of the sickle cell disease in primary hematopoietic stem cells |
DeWitt, Mark A, et al. (2016) |
| Exon Snipping in Duchenne Muscular Dystrophy in a mouse model, muscle stem cells and in human hiPSC-derived muscle cells |
Nelson, Christopher E, et al. (2016) |
| Tabebordbar, Mohammadsharif, et al. (2016) |
| Young, Courtney S, et al. (2016) |
| Young, Courtney S, et al. (2017) |
| Targeting FGFR3 oncogenic fusions for urothelial carcinomas in cell lines |
Faltas, Bishoy M, et al. (2017) |
Table 1: The main recent advances and applications of CRISPR/Cas systems.
Technical Issues
Because of the need in some instances to repeat administration of such therapy to sustain the therapeutic effect, self-limiting proinflammatory response might be triggered. An example of this is gene-editing therapy used in the treatment of Human Immunodeficiency Virus [50]. Moreover, neutralizing antibodies may be produced which annul the circulating viral vectors [17,51]. In order to minimize the toxicity and evade the immunogenicity and neutralizing antibodies produced, procedures such as delivery of carefullymeasured and smaller AAV vectors and/or ex vivo treatment of cells followed by transplanting them into the patient’s tissues can be used [52]. Nevertheless, the small adeno-associated virus (AAV) vectors affect their efficacy. Thus, there is a need for research into the development of safer and more effective non-viral-based approaches.
Since delivering a CRISPR/Cas system to T-cells is quite difficult due to the high efficiency and reliability of DNA repair pathways in human primary cells, scientists have overcome this in progenitor and hematopoietic stem cells by modifying the guide RNA chemically to increase the transfection and by delivering the Cas9 protein itself or its mRNA instead of DNA [53]. For more precise and efficient genome editing, CRISPR/ Cas9 activity could be adjusted by enhancing or inhibiting DNA repair systems. Because of the competition between NHEJ and HDR repair pathways, enhancing HDR-mediated genome editing could be achieved by inhibiting NHEJ using a specific DNA ligase inhibitor [54]. Moreover, inhibiting HDR by using novel small molecules or chemical compounds can induce NHEJ-mediated mutations [55,56].
Because of Cas9 requires proto-spacer adjacent motifs (PAMs) sequence on the crRNA-complementary strand to be specific and able to cleave the sequence of interest, different CRISPR systems with different PAM recognition sequences and different crRNA for different purposes are required to target regions that lack this PAM sequence [57]. Therefore, the offtarget effects should be carefully re-evaluated for each candidate system due to potential long-term and deleterious oncogenic effects on patients. As the specificity of CRISPR/ Cas9 is decided by just a 14 base pairs long sequence (the 12 base pairs of the guide RNA and the 2 or 3 base pairs of the PAM), the sequence length of unique 14 base pairs to be repeated is 268 Mb. Therefore, for big genomes, this increases the chances of off-target activities [35].
Possible Therapeutic Applications
The enzymatic machinery of CRISPR/Cas9 system could be harnessed to halt the growing resistance to antibiotics by attacking major virulence genes or removing microbial DNA sequences responsible for pathogenicity or antibacterial resistance [58,59]. This approach was used and scientists successfully sensitized Staphylococcus aureus to antibiotics and improved the therapeutic efficacy of antimicrobials against Staphylococcus aureus in vitro and in vivo. These candidate selective programmable antimicrobial CRISPR-Cas9 systems can potentially be used in the clinic and help evade the promiscuous bactericidal effect of conventional antibiotics [58,60,61].
CRISPR/Cas9 was efficiently used to cleave the genome of DNA viruses such as Epstein–Barr virus in human cells and to target herpesvirus genomes in herpesvirus induced diseases such as lymphomas and adenocarcinomas [62-67]. The RNAtargeting CRISPR/C2c2 system, which is guided by a single crRNA and has ribonuclease function, could be designed to knock down specific mRNA or cleave the single-stranded viral RNA that carries complementary protospacers, establishing CRISPR/C2c2 system as a novel RNA-targeting strategy [68,69].
Furthermore, by identifying the genes involved in drug resistance, CRISPR/Cas9 systems can be harnessed to develop gene-deactivating targeted therapeutics which can be in the treatment of cancer such as performing knockout for genes associated with drug-resistance to the BRAF protein kinase inhibitors used for the treatment of melanoma [36].
More importantly, in complex diseases instead of conducting laborious and time-consuming experiments to cause multiple mutations in animal models to refine the loci involved, we can potentially utilize CRISPR methodology even in human-derived cells to isolate genetic signals or identify various functions a particular pleiotropic gene [27]. However, programming CRISPR to cause mutations and create models for human diseases implies major risks, even in animals, due to possible errors in programming the guide RNA sequence that might theoretically enable CRISPR to target human genomes.
Ethical Issues
Although commercial companies could exclusively hold patents or intellectual property rights of each drug for each candidate gene edited, the most controversial ethical issue of CRISPR is its potential utilization in editing embryos or gametes [70]. It is considered unethical and even illegal to edit human embryos with the purpose of using them to achieve fertilization [71]. Although the embryos in the human tripronuclear zygotes editing trial were non-viable, the purpose of them was to implant them to achieve a pregnancy or in vitro fertilization and not for research purposes [49]. Editing preimplantation embryos is still a controversial issue since it can be exploited to “design” babies [72,73]. Many scientists have suggested that the gene versions that we have are not necessarily the perfect ones and started to call for rewriting even the normal genes [74]. Serious ethical issues have arisen from altering healthy embryos by affixing a gene that encodes a preferred phenotype to specific DNA elements that have the ability to be copied exponentially across the chromosomes to increase the chance of passing down this gene quickly to subsequent generations. However, such potential application could encourage research in this field. Even though the designed therapeutic effect could not be achieved in somatic cells in many cases, ethical concerns that arise from human germ-line modification or a moratorium imposed on editing human embryos for research purposes might restrict the clinical research and negatively affect studies on somatic cells [75]. It has been argued that law makers should allow experiments on the relatively easy-to-attain cells from patients to evaluate the effectiveness and possible off-target effects. Therefore, a framework should be established to enable the research to proceed by defining the ethical boundaries and eliminate the concerns regarding the germ-line editing.
Conclusion
In conclusion, the aforementioned proof-of-principle studies provide evidence that CRISPR/Cas genome-engineering systems have brought about a huge revolution in medical practice with a myriad of promising therapeutic applications for as yet untreatable disorders. However, this is a longstanding goal due to the tremendous technical and ethical challenges of applying this technology. Despite the theoretical benefits of the pre-implantation treatment to eradicate intractable genetic disorders before birth, it is still widely considered unethical and has negatively affected somatic cell genome-editing as well [72,76]. Unlike somatic cell genome editing, the permanent heritable changes that result from modification of sperm or egg-producing cells could be evolutionarily inappropriate and might have long-term and deleterious consequences for human genetic adaptation to environmental changes and for the whole ecosystem. Finally, germ-line editing could be applied to non-medical uses such as genetic enhancement, so the ethical frontiers that regulate the utilization of CRISPR/Cas9 technology remain to be fully addressed.
References
- https://www.technologyreview.com/s/524451/genome-surgery.
- Makarova KS, Wolf YI, Koonin EV (2013) Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res 41: 4360-4377.
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551-2561.
- Sternberg SH, Haurwitz RE, Doudna JA (2012) Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. Rna 18: 661-672.
- Ledford H (2015) CRISPR, the disruptor. Nature 522: 20-25.
- Williams BO, Warman ML (2017) CRISPR/CAS9 Technologies. J Bone Miner Res 32: 883-888.
- Erard N, Knott SR, Hannon GJ (2017) A CRISPR resource for individual, combinatorial, or multiplexed gene knockout. Molecular Cell 67: 348-354.
- Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences 109: 2579-2586.
- Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E (2016) The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532: 517-521.
- Chylinski K, Makarova KS, Charpentier E, Koonin EV (2014) Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42: 6091-6105.
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, et al. (2015) Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519: 199-202.
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, et al. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67-71.
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, et al. (2016) in vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Sci 351: 403-407.
- Chen J, Lai Y, Wang L, Zhai S, Zou G, et al. (2017) CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus Beauveria bassiana. Sci Rep p. 8.
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, et al. (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961-971.
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24: 142-153.
- Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262-1278.
- Yin H, Kauffman KJ, Anderson DG (2017) Delivery technologies for genome editing. Nat Rev Drug Discov 16: 387-399.
- Quirin KA, Kwon JJ, Alioufi A, Factora T, Temm CJ, et al. (2017) Safety and efficacy of AAV retrograde pancreatic ductal gene delivery in normal and pancreatic cancer mice. Molecular Therapy-Methods & Clinical Development.
- Mout R, Ray M, Lee YW, Scaletti F, Rotello VM (2017) in vivo delivery of CRISPR/Cas9 for therapeutic gene editing: Progress and challenges. Bioconjugate Chemistry 28: 880-884.
- Preston SP, Pellegrini M, Ebert G (2016) Hydrodynamic injection as a method of gene delivery in mice: A model of chronic hepatitis B virus infection. Programmed cell death: Methods and Protocols 1419: 109-115.
- Yu Q, Tan RZ, Gan Q, Zhong X, Wang YQ, et al. (2017) A novel rat model of nonalcoholic fatty liver disease constructed through CRISPR/Cas-based hydrodynamic injection. Mol Biotech 10: 1-9.
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, et al. (2014) Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nature biotech 32: 551-553.
- Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, et al. (2015) Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proceedings of the National Academy of Sciences 112: 10437-10442.
- Xu L, Park KH, Zhao L, Xu J, El-Refaey M, et al. (2016) CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther 24: 564-569.
- Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, et al. (2016) CRISPR/Cas9 ß-globin gene targeting in human haematopoietic stem cells. Nature 539: 384-389.
- González F, Zhu Z, Shi ZD, Lelli K, Verma N, et al. (2014) An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell stem cell 15: 215-226.
- McQueen C, Pownall ME (2017) An analysis of MyoD-dependent transcription using CRISPR/Cas9 gene targeting in Xenopus tropicalis embryos. Mechanisms of Development.
- Fichtner F, Castellanos RU, Ulker B (2014) Precision genetic modifications: A new era in molecular biology and crop improvement. Planta 239: 921-939.
- Puchta H (2016) Using CRISPR/Cas in three dimensions: towards synthetic plant genomes, transcriptomes and epigenomes. Plant J 87: 5-15.
- Choi PS, Meyerson M (2014) Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun 5: 3728.
- Bak RO, Porteus MH (2017) CRISPR-mediated integration of large gene cassettes using AAV donor vectors. Cell Rep 20: 750-756.
- Mou H, Kennedy Z, Anderson DG, Yin H, Xue W (2015) Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med 7:53.
- Davidson BL, Monteys AM, McBride JL, Boudreau R (2017) Reduction of off-target RNA interference toxicity. University of Iowa Research Foundation, Assignee. United States patent.
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, et al. (2013) CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nature protocols 8: 2180-2196.
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Sci 343: 84-87.
- Xue HY, Ji LJ, Gao AM, Liu P, He JD, et al. (2016) CRISPR-Cas9 for medical genetic screens: Applications and future perspectives. J Med Genet 53: 91-97.
- Québatte M, Dehio C (2017) Systems-level interference strategies to decipher host factors involved in bacterial pathogen interaction: from RNAi to CRISPRi. Curr Opin Microbiol 39: 34-41.
- Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, et al. (2016) Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep 6: 22555.
- Zhao N, Wang G, Das AT, Berkhout B (2017) Combinatorial CRISPR-Cas9 and RNAi attack on HIV-1 DNA and RNA can lead to cross-resistance. Antimicrob Agents Chemother 11: AAC-01486.
- Kemaladewi DU, Cohn RD (2016) Exon snipping in duchenne muscular dystrophy. Trends Mol Med 22: 187-189.
- Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, et al. (2016) in vivo gene editing in dystrophic mouse muscle and muscle stem cells. Sci 351: 407-411.
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, et al. (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Sci 351: 400-403.
- Wojtal D, Kemaladewi DU, Malam Z, Abdullah S, Wong TW, et al. (2016) Spell checking nature: versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am J Human Genetics 98: 90-101.
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, et al. (2013) Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell stem cell 13: 653-658.
- Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, et al. (2012) Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nature Med 18: 618-623.
- Bednarski C, Tomczak K, Vom Hövel B, Weber WM, Cathomen T (2016) Targeted integration of a super-exon into the CFTR locus leads to functional correction of a cystic fibrosis cell line model. PloS one 11: e0161072.
- Xie F, Ye L, Chang JC, Beyer AI, Wang J, et al. (2014) Seamless gene correction of ß-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res 24: 1526-1533.
- Liang P, Xu Y, Zhang X, Ding C, Huang R, et al. (2015) CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein & Cell 6: 363-372.
- Rogers GL, Cannon PM (2017) Gene therapy approaches to human immunodeficiency virus and other infectious diseases. Hematology/Oncology Clinics of North America 31: 883-895.
- Vandendriessche T, Thorrez L, Acosta-Sanchez A, Petrus I, Wang L, et al. (2007) Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. Lentiviral vectors for hemophilia B gene therapy. J Thromb Haemost 5: 16-24.
- Mocarski E, Quake SR, Xiong XC (2017) Antiviral treatment with low immunogenicity. Inventors of Agenovir Corporation, Assignee United States patent.
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, et al. (2015) Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nature biotech 33: 985-989.
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, et al. (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nature biotech 33: 538-542.
- Yu C, Liu Y, Ma T, Liu K, Xu S, et al. (2015) Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16: 142-147.
- Du J, Shang J, Chen F, Zhang Y, Yin N, et al. (2017) A CRISPR/Cas9-based screening for non-homologous end joining inhibitors reveals ouabain and penfluridol as radiosensitizers. Mol Cancer Ther.
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733-740.
- Citorik RJ, Mimee M, Lu TK (2014) Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nature biotech 32: 1141-1145.
- Goren M, Yosef I, Qimron U (2017) Sensitizing pathogens to antibiotics using the CRISPR-Cas system. Drug Resist Updat 30: 1-6.
- Park JY, Moon BY, Park JW, Thornton JA, Park YH, et al. (2017) Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci Rep 7.
- Bikard D, Barrangou R (2017) Using CRISPR-Cas systems as antimicrobials. Curr Opin Microbiol 37: 155-160.
- Yuen KS, Chan CP, Wong NH, Ho CH, Ho TH, et al. (2015) CRISPR/Cas9-mediated genome editing of Epstein–Barr virus in human cells. J Gen Virol 96: 626-636.
- Diemen FR, Lebbink RJ (2017) CRISPR/Cas9, a powerful tool to target human herpesviruses. Cellular Microbiol p. 19.
- Bi Y, Sun L, Gao D, Ding C, Li Z, et al. (2014) High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS pathogens 10: 1004090.
- Xu X, Fan S, Zhou J, Zhang Y, Che Y, et al. (2016) The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of a-4 gene transcription. Virology Journal 13: 152.
- Van Diemen FR, Kruse EM, Hooykaas MJ, Bruggeling CE, Schurch AC, et al. (2016) CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS pathogens 12: 1005701.
- Roehm PC, Shekarabi M, Wollebo HS, Bellizzi A, He L, et al. (2016) Inhibition of HSV-1 replication by gene editing strategy. Sci Rep 6: 23146.
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, et al. (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353: 5573.
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, et al. (2015) Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Molecular cell 60: 385-397.
- Segers S, Mertes H, Pennings G (2017) Stem cell derived gametes: A slippery slope towards designer babies?. In: Annual Meeting ESHRE 32: 97.
- De Araujo M (2017) Editing the genome of human beings: CRISPR-Cas9 and the ethics of genetic enhancement. J Evol Technol 27: 24-42.
- Lanphier E, Urnov F (2015) Don't edit the human germ line. Nature 519: 410-412.
- Lee MJ, Sophia S (2017) Arguments for and against germline intervention: A critical review of Ronald Green’s babies by design. Journal of Healthcare Ethics and Administration p. 3.
- https://www.technologyreview.com/s/535661/engineering-the-perfect-baby/.
- Mulvihill JJ, Capps B, Joly Y, Lysaght T, Zwart HA, et al. (2017) Ethical issues of CRISPR technology and gene editing through the lens of solidarity. British Medical Bulletin 122: 17-29.
- Cyranoski D, Reardon S (2015) Embryo editing sparks epic debate. Nature 520: 593-594.



