- (2000) Volume 1, Issue 2
Kevin Sargen1,2, Andrew G. Demaine1, Andrew N. Kingsnorth2*
1Departments of Molecular Medicine and 2Surgery Plymouth Postgraduate Medical School, Derriford Hospital. Plymouth, United Kingdom.
Received March 15th, 2000, Accepted May 12th, 2000
Context Pro-inflammatory and regulatory cytokines play a key role in the pathogenesis of acute pancreatitis. Genetic loci encoding cytokines have been shown to be polymorphic, in some cases influencing protein expression.
Objective To investigate if TNF and IL-10 gene loci are associated with the occurrence or severity of acute pancreatitis.
Setting Acute surgical unit within large district hospital serving a population of 500,000.
Methods Three TNF microsatellite loci (TNFa, TNFb, TNFc), the TNF-308 polymorphism, the IL-10. G microsatellite locus, and 3 bi-allelic polymorphisms in the IL-10 5’ region were typed using PCR in 135 acute pancreatitis patients and ethnically matched normal controls (n=107). Aetiology of disease was determined and patients grouped according to disease severity by assigning an organ failure score or classification according to the Atlanta system.
Main outcome measures Allelic frequency of polymorphic loci in patients with different aetiology and disease course in acute pancreatitis.
Results No difference was noted in allelic frequency of any of the cytokine gene loci between groups stratified according to disease severity. When aetiology was studied again there was no significant difference in allelic frequency.
Conclusions The cytokine gene polymorphisms studied play no part in determination of disease severity or susceptibility to acute pancreatitis.
Inflammatory Response; Interleukin-10; Tumour Necrosis Factor
bp: base pair; ERCP: endoscopic retrograde cholangiopancreatography; IL: interleukin; Kb: kilobase; OFS: organ failure score(s); MHC: major histocompatibility complex; TMC: tumour necrosis factor
The clinical course of acute pancreatitis is often mild with only minimal associated organ dysfunction, but a significant proportion of patients develop severe pancreatitis which is associated with organ failure, systemic complications, local inflammatory and infective manifestations and, in up to 10% of cases, death.
Pro-inflammatory and regulatory cytokines play a fundamental role in the local and systemic inflammatory response in the initial stages of disease and in the development of severe acute pancreatitis [1, 2]. Tumour necrosis factor-a (TNF-a) and TNF-b are potent mediators of the immune response and are central to the initiation of the cytokine network, causing other proinflammatory cytokines such as interleukins- 6 and 8 (IL-6, IL-8) to be produced [3, 4]. TNF expression in the pancreas is increased by the onset of experimental pancreatitis [5- 7], and antagonism of TNF reduces the severity of local pancreatic inflammation [5, 8]. Levels of soluble TNF receptors, indicators of TNF activity, have been found to be increased in patients with severe disease [9], and TNF blockade has been shown to reduce mortality and ameliorate markers of severe systemic disease in experimental acute pancreatitis [10].
Interleukin–10 (IL-10) is a cytokine with regulatory, anti-inflammatory effects. For example, macrophage secretion of TNF and other pro-inflammatory cytokines are profoundly suppressed by IL-10 [11]. IL-10 administration is known to attenuate local pancreatic inflammation in experimental models of acute pancreatitis [12, 13], and also reduces mortality rates in experimental animals [14]. In patients with acute pancreatitis, IL-10 serum levels are reduced in severe as opposed to mild disease [15].
The TNFa and b loci are located within the major histocompatibility complex (MHC), tandemly arranged over 7 kilobases (Kb), within the class III region. There are a number of polymorphic sites within the TNF loci, including an AC/GT repeat (TNFa), two closely linked TC/GA repeats (TNFb and TNFc) [16], and a bi-allelic polymorphism consisting of a G to A substitution at position -308 in the TNF-a promoter region [17]. The TNFa and TNFb microsatellites are 3.5 Kb upstream of the TNF-b locus, TNF-c being located in the first intron.
Whilst there is no conclusive evidence regarding the functional role of the -308 polymorphism, alleles a2 and c2 of the TNFa and TNFc microsatellites have been associated with higher TNF production [18], as have certain TNF haplotypes [19]. Certain TNF alleles and haplotypes have been found to be associated with inflammatory diseases such as Crohn’s disease [20].
The gene encoding IL-10 has been mapped to chromosome 1 [21]. The IL-10 promoter contains numerous and varied transcription factor binding sites [22], and two regions within it, -1100 to –900, and -800 to –300, have been identified as having effects upon gene transcription [23]. Close to or within this region are 4 polymorphic sites, 3 single base transitions at –1117 (G to A substitution), -854 (C to T substitution), and –627 (C to A substitution). There is also a (CA)n repeat, the IL10.G microsatellite, located between positions -1193 and –1151 [24]. It is postulated that polymorphisms in this region may be functional through their associated alteration of an important transcription factor binding site. Indeed, the slightly more common allele of the –1117 polymorphism (-1117.G) has been shown to produce higher in vitro production of IL-10 by stimulated monocytes [25].
In this study we have evaluated the association of the TNF and IL-10 genes with acute pancreatitis and resultant disease severity.
Patients
One hundred and 35 Caucasian patients admitted with a diagnosis of acute pancreatitis were identified shortly after admission to hospital. The criteria for diagnosis of acute pancreatitis were: a clinical history consistent with the disease, appropriate radiological evidence, and a serum amylase level greater than 660 U/L (Hitachi 911, Hitachi Corporation, Japan; normal range <220 U/L). The progress of individuals with regard to development of complications was monitored during their disease episode, enabling patients to be classified as having mild disease or severe local or systemic disease according to criteria defined by the Atlanta convention [26]. Maximal organ failure scores (OFS) [27] were also calculated to provide another means of patient stratification.
Aetiology of acute pancreatitis was classified as being due to gallstones in the presence of appropriate radiological or endoscopic retrograde cholangiopancreatography (ERCP) findings (n=77), alcoholic if subjects admitted on questioning to being consistent heavy consumers of alcohol (n=21), or idiopathic if no other identifiable cause could be discovered (n=37). Consistent heavy consumption of alcohol was defined as daily consumption of greater than 80 g of alcohol per day over a time period exceeding six months. No subjects who had acute pancreatitis secondary to excessive ‘binge’ consumption of alcohol were encountered during recruitment for this study. Any history of alcohol consumption recorded in medical notes was confirmed by interviews with individual patients.
Patients who had clinical, radiological, or ERCP evidence suggestive of a diagnosis of chronic pancreatitis were excluded. CT or ERCP was performed if there was a clinical suspicion of chronic pancreatitis in those patients with alcohol induced disease. In 15 of 21 patients (71%) classified as having acute alcohol-induced acute pancreatitis CT or ERCP was performed. The other 28.6% (6 of 21) who had no clinical suspicion of chronic pancreatitis had an adequate ultrasound scan (pancreas clearly visualised), with no evidence of chronic pancreatitis in this imaging modality.
Controls
One hundred and seven Caucasoid cord blood samples following a normal healthy obstetric delivery were used to obtain control allele and genotype frequencies in the TNF assays, but up to 136 controls were used in the IL-10 assays.
DNA Preparation
High molecular weight DNA was prepared from peripheral venous blood using the Nucleon II DNA extraction kit (Scotlab, Lanarkshire, UK).
TNF Microsatellite Typing
The three microsatellite loci, TNFa, TNFb, and TNFc were amplified by the polymerase chain reaction (PCR) as a one-step procedure using oligonucleotide amplimers synthesised (Pharmacia Biotech, Sweden) from previously described sequences [16, 28]. 100-500 ng of genomic DNA was added to a final reaction volume of 20 ml with 0.5 mM of each amplimer pair (reverse amplimer 5’-end-labelled with 50,000 cpm of 32P), 10 mM Tris-HCL (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 300 mM of each dNTP, and 0.8 units of Taq DNA polymerase (HT Biotechnology, Cambridge, UK). PCR for TNFa and TNFb were performed in a Cyclogene thermocycler (Techne, Cambridge, UK) under the following conditions: 95 °C for 3 minutes, then 30 cycles of 95 °C for 1 minute, 62 °C for 1 minute, and 68 °C for 1 minute followed by a 3 minute extension at 68 °C. The TNFc microsatellite was amplified in an identical way but employing an annealing temperature of 58 °C. Amplification products (6 ml) mixed with 3 mL of Stop solution containing Formamide (Amersham Life Science, Buckinghamshire, UK) were then separated on a 6% polyacrylamide gel with 8 M urea at 1700 V for 2.5 hours (TNFa and TNFc), or 3 hours (TNFb). After drying the gels were exposed to Kodak XLS5 X-ray film (Scientific Imaging Systems, Cambridge, UK) with intensifying screens at –70 °C for 18 hours.
TNF -308 Typing
Amplification of an 836 base pair (bp) fragment of the TNF promoter region was performed in two PCR-SSP reactions. These employed a common forward amplimer:
5’-CTGCATCCCCGTCTTTCTCC-3’ and one of two reverse amplimers with a 3’ mismatch corresponding to a G or an A at position -308:
5’-ATAGGTTTTGAGGGGCATCG-3’
5’-ATAGGTTTTGAGGGGCATCA-3’
Amplimers for a 256 bp control amplicon from exon 15 of the adenomatous polyposis coli gene was included in the same reaction.100-500 ng of genomic DNA was added to a final reaction volume of 20 ml with 0.5 mM of each amplimer pair, 10 mM Tris-HCL (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 300 mM of each dNTP, and 0.8 units of Taq DNA polymerase (HT Biotechnology, Cambridge, UK). The assay was performed in a PTC-200 Thermal Cycler (MJ Research, Essex, UK) under the following cycling conditions: 96 °C for 3 minutes, then 30 cycles of 96 °C for 45 seconds, 55 °C for 80 seconds, and 72 °C for 2 minutes followed by a 3 minute extension at 72 °C.
The entire reaction volume plus 5 ml of Orange G track dye were loaded into a 1% agarose gel containing ethidium bromide. Gels were electrophoresed for 20 minutes at 200 V. The gels were then photographed under UV light (320 nm) and scored for the presence or absence of an allele specific band providing a PCR control band was present.
IL-10 –1117, -854, and –627 Typing
Amplification of an 587 bp region containing the three polymorphisms at positions –1117, -854, and –627 was performed in a Cyclogene thermocycler (Techne, Cambridge, UK) in 20 ml volumes using the forward amplimer 5’-ATCCAAGCAAC ACTACTAA-3’ and reverse amplimer 5’- TAAATATCCTCAAAGTCCC-3’. The reaction mixture contained 0.5 mM of each amplimer pair, 10 mM Tris-HCL (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X- 100, 300 mM of each dNTP, and 0.8 units of Taq DNA polymerase (HT Biotechnology, Cambridge, UK). PCR product was blotted onto Hybond N+ nylon transfer membrane (Amersham, Bucks, UK).
Two 5’-end labelled digoxigenin probes were used to detect each polymorphism. For the - 1082 locus, the probes were TAAGGCTTCTTTGGGAGG and TAAGGCTTCTTTGGGAAG (wash T° = 60 °C). For the -819 locus the probes were CAGGTGATGTAACATCTCTGTCG and GCACAGAGATATTACATCACCTGT (wash T° = 70 °C). For the -592 locus the probes used were TGTGACCCCGCCTGCC and CTGTGACCCCGCCTGAC (wash T° = 62 °C). Probes were hybridised at 42 °C and washed in 3.2 M tetra-methyl-ammonium chloride (TMAC) solution. Anti-digoxigenin antibody was used in conjunction with a chemoluminescent detection system to detect bound probe.
IL-10.G Microsatellite Typing
Amplification of a region containing the microsatellite was performed using the forward amplimer 5’- TCCTTCCCCAGGTAGAGCAACACTCC- 3’, and reverse amplimer 5’- TCCCAAAGAAGCCT TAGT AGTGTTG- 3’. The former was 5’-end labelled with [g 32P] ATP. The reaction mixture contained 0.5 mM of each amplimer pair, 10 mM Tris- HCL (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton X-100, 300 mM of each dNTP, and 0.8 units of Taq DNA polymerase (HT Biotechnology, Cambridge, UK).
Amplification products (6ml) mixed with 3 ml of Stop solution containing Formamide (Amersham Life Science, Buckinghamshire, UK) were then separated on a 6% polyacrylamide gel with 8 M urea at 1700 V for 2.5 hours, and alleles revealed with autoradiography.
Local ethical committee approval had been obtained, confirming the study protocol conformed to the guidelines of the Declaration of Helsinki (1975, revised 1983) and patients gave informed consent.
Patient groups were compared with respect to age, which is expressed as median and range. Distribution of age was tested for normality using Statgraphics software (Statistical Graphics Corp. USA. 1996). Distribution in subject groups was tested for normality using the c2 goodness-of-fit statistic, the Shapiro-Wilkes W statistic, the z-score for skewness, and the z-score for kurtosis. As no group fitted normal distribution this tends to invalidate comparison of means and standard deviations using conventional testing. Therefore, a nonparametric test (Kolmogorov-Smirnov) was used to compare whether there is a statistically significant difference in distribution of age between study groups.
Comparisons of allelic and haplotype frequencies between patient subgroups and controls were made in 2 x 2 contingency tables using the c2 method. Corrections were made if necessary for small sample numbers using Fisher’s exact test. P values were corrected for multiple comparisons made using the Bonferroni inequality method. Corrected p values of less than 0.05 were considered statistically significant.
Patient Groups
Of the 135 patients in this study, 97 had mild disease as defined by the Atlanta criteria [26]. Of the 38 patients with severe disease, 30 had systemic complications defined by at least one organ dysfunction, 21 patients had a local pancreatic collection (fluid, necrosis, or pseudocyst), of whom only 8 did not have concurrent systemic complications. 18 patients had organ failure scores of 3 or more, with the respiratory system accounting for at least 3 of the score [27].
In 21 patients the episode of acute pancreatitis was caused by alcohol (³80 g/day), in 77 the cause was cholelithiasis, and in 37 patients no cause could be identified (classified as idiopathic).
Age and sex characteristics of controls and patient subgroups are displayed in Table 1. Testing with a non-parametric test revealed that distribution of age was younger in patients with mild acute pancreatitis compared to those with severe disease (p=0.007, Kolmogorov-Smirnov test), but not those with an OFS³3 (p=0.11, Kolmogorov-Smirnov test). The group with alcohol induced acute pancreatitis were significantly younger than those with another aetiology (p<0.001, Kolmogorov-Smirnov test).
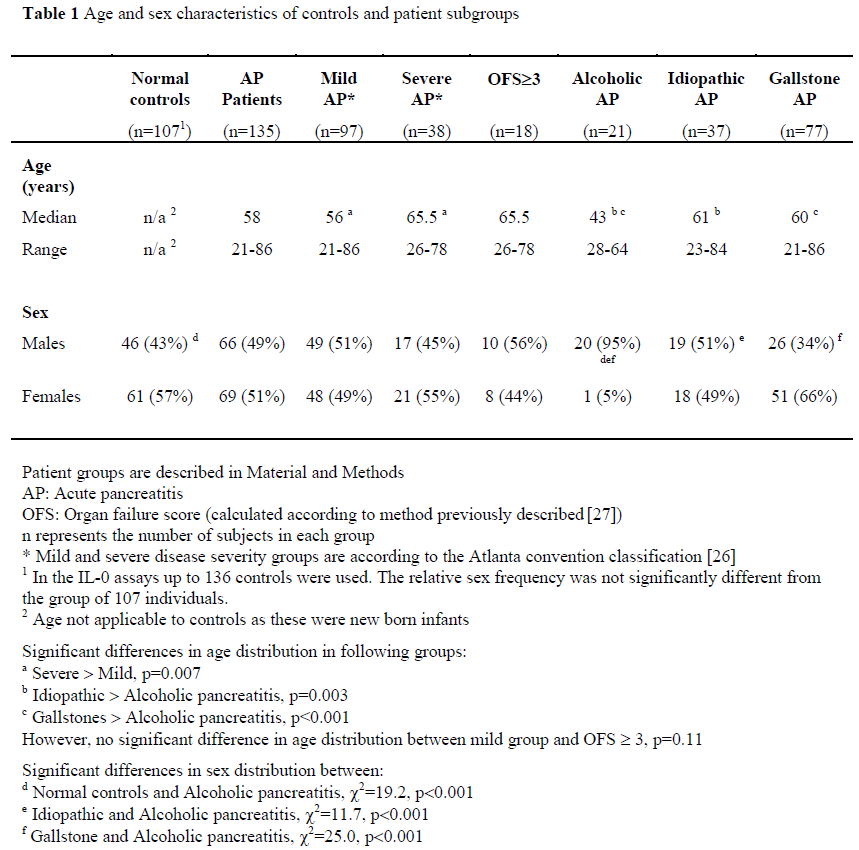
Sex distribution was comparable between all groups, apart from the group with alcoholic pancreatitis, where all but one subject was male (normal controls vs. alcoholic pancreatitis, c2=19.2, p<0.001, Idiopathic vs. alcoholic pancreatitis, c2=11.7, p<0.001, Gallstone vs. alcoholic pancreatitis, c2=25.0, p<0.001).
TNF Microsatellite Allele Frequencies
Allelic frequencies were measured in 100% of patients. We observed 13 of 14 known alleles of TNFa in the population studied. Six of the seven known alleles of TNFb were found in our patient and control groups. TNFc is biallelic. Table 2 shows the allelic frequencies at the three loci in control and subject groups.
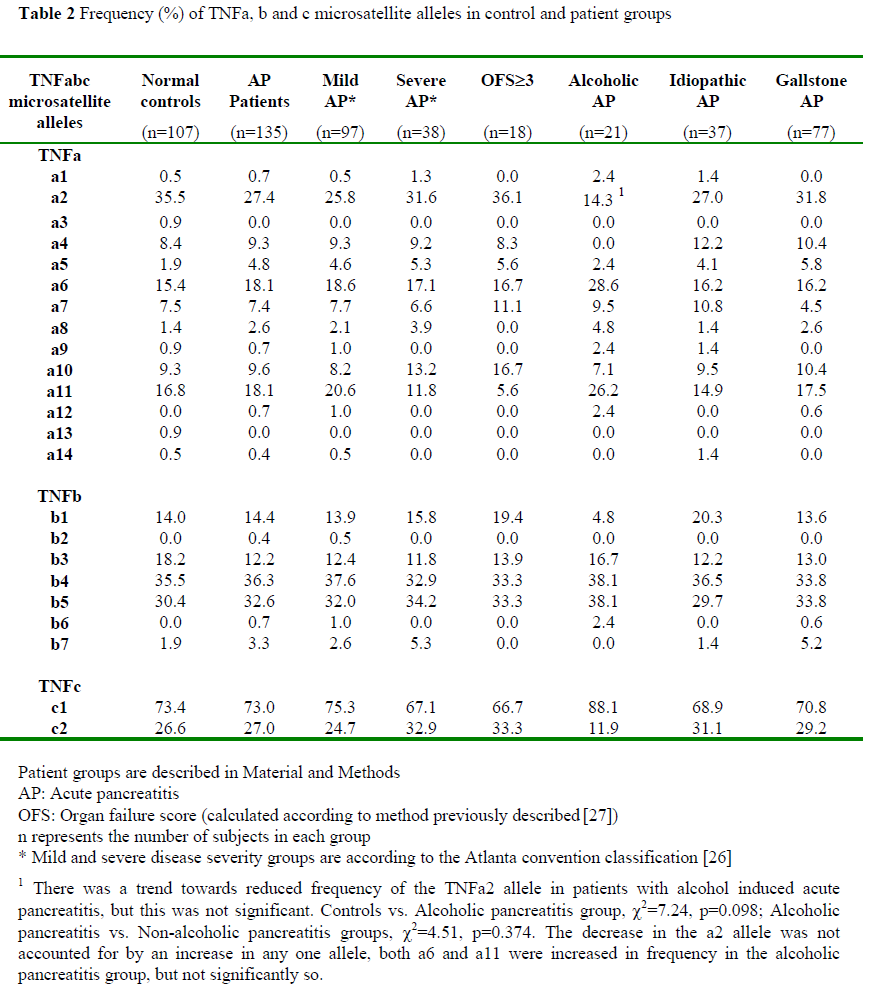
When patients were grouped according to aetiology there was a trend towards reduced frequency of the TNFa2 allele in those patients with alcoholic acute pancreatitis compared to controls (c2=7.24, p=0.098), or compared to those with pancreatitis of other aetiology (c2=4.51, p=0.374).
There are no statistically significant differences in the allelic frequencies of the TNF-a, TNF-b, or TNF-c loci when comparing all acute pancreatitis patients with controls or when patients grouped according to disease course (mild vs. severe, mild vs. OFS³3) were compared.
TNF Microsatellite 3 Locus Haplotype
Haplotypes, alleles combining in non-random association across the TNF 3 locus microsatellite system, have been characterised by maximum likelihood estimates in previous population based and family studies [16, 28, 29] and their existence has been confirmed in homozygous cell lines [30]. TNF haplotype analysis was performed using all 11 haplotypes described in European populations [29, 31]. There were no significant differences in haplotype frequencies between those groups with mild acute pancreatitis, severe acute pancreatitis, or organ failure, or between any of the patient groups and the normal healthy controls (data not shown).
When patients were stratified according to disease aetiology, the haplotype TNFa2b1c2 showed a trend toward reduced frequency in the group with alcohol induced acute pancreatitis. TNFa2b1c2 was present in 4.8% of individuals compared with 24.3% of the controls (c2=4.03, p=0.495), 31.9% of patients with non-alcoholic pancreatitis (c2=6.76, p=0.108). As can be seen, none was significant after correction for multiple comparisons.
There was no difference in any other haplotype frequency between the groups.
-308 Polymorphism
Allelic frequencies of this bi-allelic polymorphism were measured in 97% of patients. There was no difference in allelic frequency between the normal controls and patients or between the different patient groups. TNF -308.G had a frequency of 78.3% in normal controls compared to 84.4% in all acute pancreatitis patients, whilst the frequency of TNF-308.A was 21.7% in controls and 15.6% in patients (c2=2.78, p=0.1).
IL-10 G Microsatelite
Allelic frequencies were measured in 100% of patients and are shown in Table 3. We observed 11 of 15 alleles of the IL-10.G microsatellite.
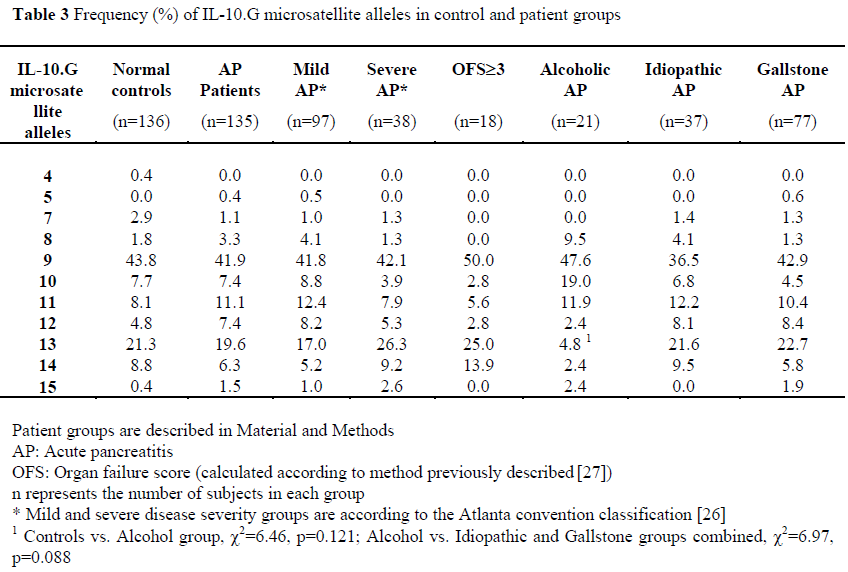
There were no differences in allele frequencies between patients and controls, or between controls and patient groups stratified according to disease severity (mild vs. severe, mild vs. OFS³3).
When patients were stratified according to aetiology of acute pancreatitis, although there was a reduction in the frequency of the IL-10.G13 allele in patients with alcoholic pancreatitis compared to normal controls (4.8% vs 21.3%, c2=6.46, p=0.121), or compared to those with non-alcoholic pancreatitis (4.8% vs. 22.2%, c2=6.97, p=0.088); none of these was significant after correction for multiple comparisons.
IL-10 Bi-Allelic Polymorphisms
In both controls and patient groups observed gene frequencies did not differ significantly from expected so Hardy-Weinberg equilibrium was established. Due to the presence of large numbers of heterozygotes at each of the 3 loci, haplotypes across the 3 loci were only able to be assigned in 68 of 127 controls (53.5%) and 71 of 124 patients (57.3%). Therefore analysis involving haplotypes was not helpful.
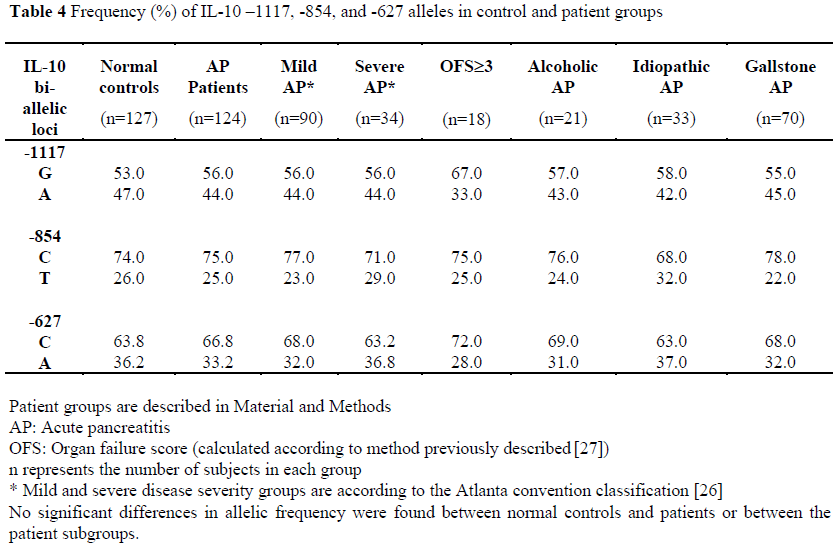
Allelic frequencies of the –1117, -854, and – 627 loci were measured in 92% of patients and are displayed in Table 4. There was no significant difference between the normal controls and patients or between the different patient groups.
This study characterises cytokine gene polymorphisms in patients with acute pancreatitis. We have shown that the TNF and IL-10 gene polymorphisms studied are not associated with disease severity or susceptibility in acute pancreatitis.
The importance of both pro-inflammatory and regulatory cytokines in the pathogenesis of acute pancreatitis is well established. Animal models have shown that TNF plays an important role in the early disease pathogenesis. Increased TNF expression has been demonstrated in these animal models [5-7], and TNF antagonism is known to ameliorate local and systemic markers of disease [5, 8, 10]. Since TNF production in individuals has been shown to be genetically influenced [18], we hypothesised that polymorphism of the TNF gene locus may influence disease susceptibility and course in acute pancreatitis.
This study of the 3-locus TNFabc microsatellites in a group of subjects with acute pancreatitis shows no association between severity of disease and TNF microsatellite alleles or haplotype. The lack of an association with the -308 polymorphism is similar to the results reported in a study in patients with severe sepsis [31].
The regulatory cytokine, IL-10, is undoubtedly important in the pathogenesis of acute pancreatitis. Unlike TNF, it is antiinflammatory, and reduces macrophage secretion of TNF [11]. As a consequence, experimental data with animals suggests that IL-10 acts to suppress the inflammatory process associated with acute pancreatitis. Because a locus within the IL-10 promoter has been shown to influence IL-10 production [25], we hypothesised that IL-10 promoter region polymorphisms may influence the development or severity of acute pancreatitis.
At present the importance of the IL-10.G microsatellite in regulating IL-10 production is under investigation. However, characterisation of the IL-10 promoter region in which the IL-10 polymorphisms described in this study occur, show that there are many transcription sites close to these polymorphisms, which may be influenced by them. For example, there are two potential NFkB/REL sites 60 and 80bp immediately upstream of IL10.G [22], which may be involved in cytokine mediated modulation of IL-10 expression, so influencing the pathogenesis of inflammatory disorders such as human acute pancreatitis.
We have shown that the bi-allelic polymorphisms of the IL-10 promoter have no association with susceptibility or severity of acute pancreatitis. We also investigated the potentially more informative microsatellite, IL-10.G, and again can show no influence upon severity or susceptibility to acute pancreatitis.
Overall, because of the failure of any differences we noted in frequencies of TNF and IL-10 polymorphisms between study groups to achieve statistical significance, we have demonstrated no association between TNF and IL-10 gene polymorphisms severity or susceptibility to acute pancreatitis. On the basis of our group of patients, it appears that the loci, or at least their markers that we have studied, are not associated with acute pancreatitis. However, as we noted a trend toward reduced frequency of the TNFa2 allele in patients with alcohol induced pancreatitis, we are expanding studies to determine frequency of this marker in a larger group of patients with alcohol induced acute and chronic pancreatitis, as we believe this is worthy of further study.
This work was supported by a research grant from Plymouth Hospitals NHS Trust