Review Article - (2016) Volume 1, Issue 2
Chakraborty AK*, Roy T and Mondal S
Department of Biotechnology and Biochemistry, Oriental Institute of Science and Technology (OIST), Vidyasagar University, West Bengal, India
*Corresponding Author:
Dr. Asit Kumar Chakraborty
Department of Biotechnology and Biochemistry
Oriental Institute of Science and Technology (OIST)
Vidyasagar University, Medinipur
West Bengal 721102, India
Tel: 919339609268
E-mail: chakraakc@gmail.com
Received date: October 25, 2016; Accepted date: November 16, 2016; Published date: November 21, 2016
Citation: Chakraborty AK, Roy T, Mondal S. (2016) Development of DNA nanotechnology and uses in molecular medicine and biologyy. Insights in Biomed. 2016, 1:2.
DNA is hereditary material with simple and stable building blocks of phosphate- 2’deoxy ribose-organic nitrogenous base with unique 3-D structure with AT-GC paring of two anti-parallel strands. DNA in solid or solution is a good carrier of electrons and biocompatable with binding efficiency to many proteins, organic molecules and metal ions. With the advent of nanotechnology in silver-gold nanoparticles and carbon nanoparticles, DNA has now been utilized as good source of nanotechnology material. In DNA nanotechnology, Watson-Crick DNA molecules are arranged into variety of nanostructures in the range of 10-100 nm size under special physical conditions due to electrostatic attraction among free electrons of sugar and phosphate oxygen and base nitrogen. However, different cohesive or sticky ends or loop structures among oligonucleotide staple strands have helped to make 3-D DNA nanostructures with various shapes. Holiday junction formation during crossover of chromosomes is the basis of DNA nanotechnology as millions fold compacted 3-D DNA structure is inherited in DNA itself. DNA tiles are hydrogen bonded few oligonucleotides which have cross sharing among themselves at the both ends. DNA-origami is principle when one big single stranded circular DNA crossovers with hundreds of short antisense oligonucleotide staple strands at different positions giving different shapes. In principle, when DNA tiles or DNA origami are allowed to crystal formation at 90°C to 4°C transition in presence of 10-20 mM MgCl2, DNA nanocrystals are formed. Thus, gold and silver as well as many drugs were impregnated in the DNA nanocages that were targeted to many pathogens and cancer cells. Such co-crystallised nano-drug delivery system also has been integrated to antisense/ribozyme/dicer molecular medicine. Recently, solid DNA nanotechnology applications combining DNA with proteins or cellulose and cross-linked with streptavidin-biotin are used in nanochip, nanosensor and nano-robotic technologies.
Keywords
DNA tiles; DNA nanotubes; DNA origami; DNA-nanocages; Drug delivery
Introduction
The term "nanotechnology" was first defined by Dr. Norio Taniguchi in 1974 as process and utilization of materials made of single atom or molecule [1]. The nanomaterial exists in a very small scale (10-6 of a millimetre) as exemplified, a human hair is about 80000 nanometers and our hereditary DNA molecule (chromosome) is about only 5-10 nanometers. Nano-particles or nano-medicines are 100-10000 times smaller than a human cell (~20-50 μm) and much smaller than microorganisms (~1-5 μm) that to be targeted or to be modified (Figure 1).
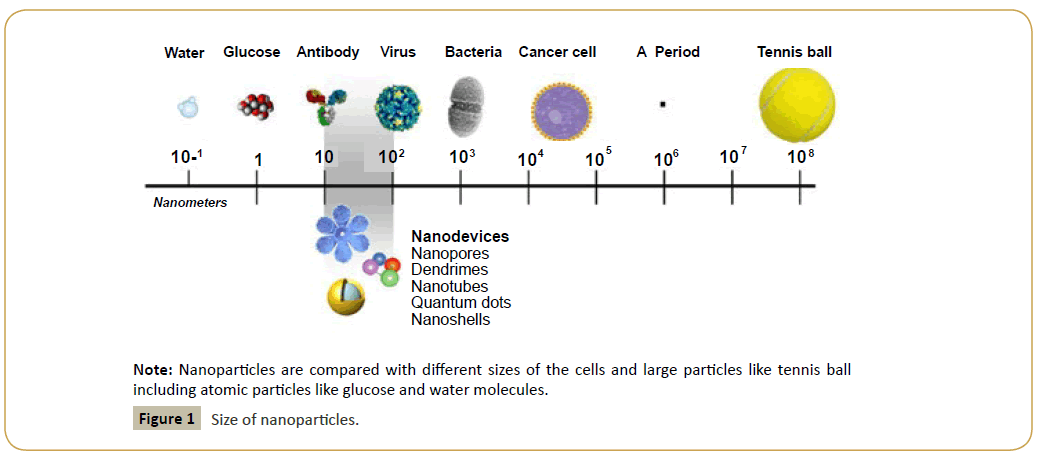
Figure 1: Size of nanoparticles.
Note: Nanoparticles are compared with different sizes of the cells and large particles like tennis ball including atomic particles like glucose and water molecules.
DNA nanotechnology was started due to the development of Cluster science and Scanning tunneling microscope followed by discovery of fullerenes in 1985 that had demonstrated how wonder properties of single cell carbon film (200x strength than steel) could exist in nature [2]. Nano-materials prepared from transition metals like silver or gold were complexed with drugs that eradicated many untreatable MDR diseases and metastasis cancer in human. The basis for nano-medicine approaches were pioneered by the earlier discoveries: (i) liposomes in drug delivery and in DNA transfection, (ii) controlled release system of macromolecules [3], (iii) circulating stealth polymeric nanoparticles [4], (iv) quantum dot bio-conjugate system [5] and (v) nanowire nano-sensor dates [6].
Success of Silver-Gold Nano-Particles and Graphenes
Most published nano-particles today are silver and gold nanoparticles. Silver (Ag) is a transition metal element having atomic number-47 (1s2, 2s2, 2p6, 3s2, 3p6, 3d10, 4s2, 4p6, 4d10, 5s) and atomic mass-107.87 whereas gold (Au) has an atomic number-79 (Ag.5s2, 5p6, 5d10, 6s) and atomic mass 170.23. Inert gas condensation or co-condensation techniques are used where evaporation of metal into an inert atmosphere with the subsequent cooling for the nucleation and growth of the nanoparticles are done. For the biosynthesis silver/gold nanoparticles, 1.5 ml of plant extract was mixed with 30 ml of 1 mM AgNO3 or 1 mM HAuCl2 solution and was incubated at 28C for 24 hrs until colourless silver nitrate solution turned brown colour or pale yellowish HAuCl2 [7]. The mixture was centrifuged at 6000rpm for 10 min and the pellet was re-suspended in sterilized double distilled water and then sprayed on glass slide to make thin film. The thin film was kept in hot air oven to dry and then the thin film was used for the TEM/SEM analysis (Figure 2). The antibacterial efficiency of the nano-particles was determined by introducing the particles into a media containing bacteria in petridisc. The nanoparticles were found to be completely cytotoxic to bacteria like Escherichia coli and Pseudomonas aeruginosa for surface concentrations as low as 8 μg of Ag/cm2 [8-10].
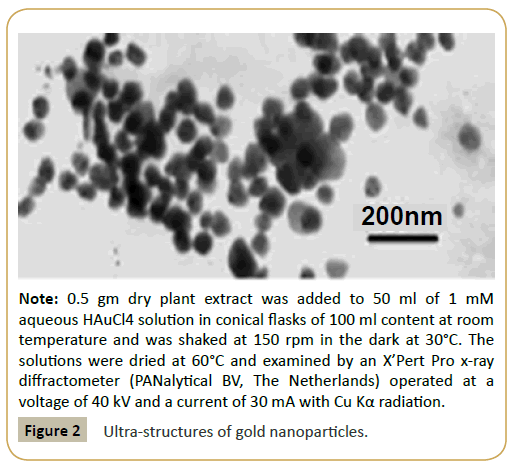
Figure 2: Ultra-structures of gold nanoparticles.
Note: 0.5 gm dry plant extract was added to 50 ml of 1 mM aqueous HAuCl4 solution in conical flasks of 100 ml content at room temperature and was shaked at 150 rpm in the dark at 30°C. The solutions were dried at 60°C and examined by an X’Pert Pro x-ray diffractometer (PANalytical BV, The Netherlands) operated at a voltage of 40 kV and a current of 30 mA with Cu Kα radiation.
DNA Nano-Technology
DNA is a chain of phosphate-pentose sugar which attached to different organic bases (adenine, cytosine, guanine and thymidine) and ds-DNA (genetic material in living cells) means two monomeric chains of DNA intertwined in opposite directions in Watson-Crick Model (Figure 3). Solid DNA is thread like whitish stable structure which is very soluble in water giving viscous opaque solution and could be detected in ng quantities by fluorescence banding of ethidium bromide stained agarose gel or 260 nm UV spectrometry. Biological synthesis of DNA by enzyme, DNA polymerase or in vitro synthesis of DNA by automated oligo-synthesizer is easy task. Similarly, topological inter-conversions of circular DNA molecules (plasmids) by enzyme DNA topoisomerases are now easy to see the inter-conversion of coiled-relaxed, monomer-catenenes, knotted-unknotted DNA topological isomers. Interestingly, DNA ligase could arrange head to tail joining of small ds-DNA molecules into massive DNA structure as seen in the chromosomes 2000000-20000000 [11,12]. In nature, similar chain structure also have been found in other systems like (i) cellulose is chains of hexose sugar (β1- >4 linkage) and in starch (α1->4 linkage),, (ii) proteins are chains (up to few thousands) of 20 different amino acids joined by –CO-NH- peptide bonds and (iii) in RNA which is very similar to monomeric DNA except in place of thymidine usually uracil present. Important thing, such chains could form complex 2-D and 3-D structures naturally and their roles in biology inevitably were emerging out recently. ds-DNA presents in bacterial genome (usually 1-2 molecules in bacteria, few in fungi and yeast and more than twenty in parasites and 46 in human diploid cells). The bacterial chromosome contains 3000000-5000000 bases but circular plasmids exist in many copies as only few thousand bases carrying drug resistant genes (amp, tet, neo, str, cat) but could be larger as conjugative MDR plasmids (~100-500 kb) containing 10- 15 mdr genes and few dozen of Tra and Tnp genes [13,14]. Such DNA could form topological isomers with difference in linking number or form knot and catenenes as exemplified in (Figure 4) [15]. It is noteworthy to say that now all enzymes have been purified and available and you could test how such enzymes (DNA topoisomerases, DNA ligase, Taq DNA polymerase) could form complex structures from monomer substrates.
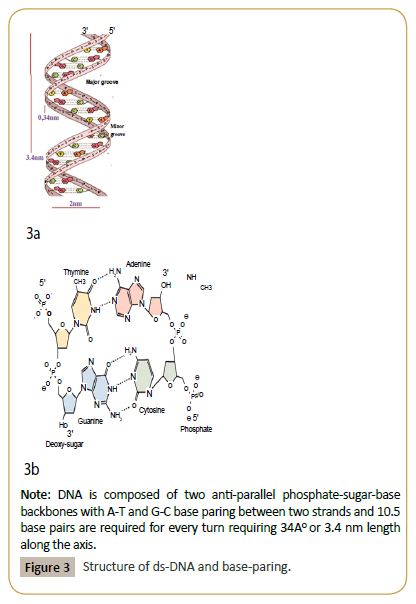
Figure 3: Structure of ds-DNA and base-paring.
Note: DNA is composed of two anti-parallel phosphate-sugar-base backbones with A-T and G-C base paring between two strands and 10.5 base pairs are required for every turn requiring 34AO or 3.4 nm length along the axis.
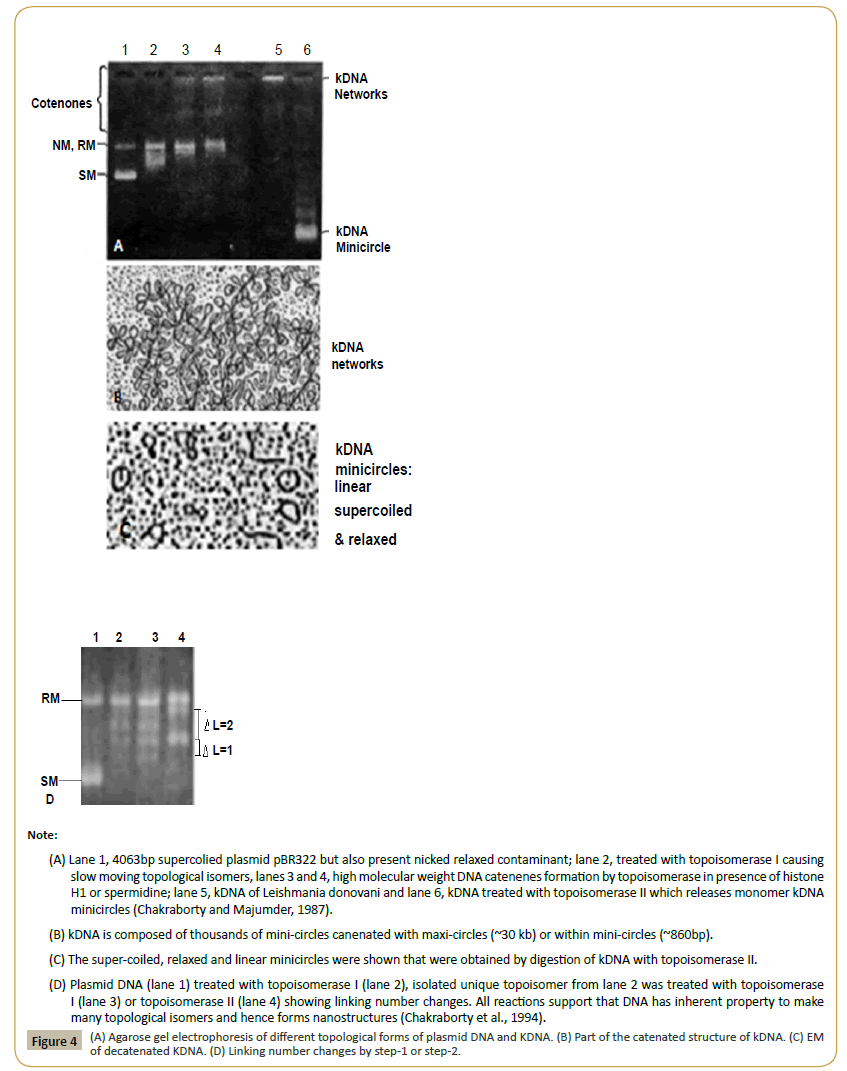
Figure 4: (A) Agarose gel electrophoresis of different topological forms of plasmid DNA and KDNA. (B) Part of the catenated structure of kDNA. (C) EM of decatenated KDNA. (D) Linking number changes by step-1 or step-2.
Note: (A) Lane 1, 4063bp supercolied plasmid pBR322 but also present nicked relaxed contaminant; lane 2, treated with topoisomerase I causing slow moving topological isomers, lanes 3 and 4, high molecular weight DNA catenenes formation by topoisomerase in presence of histone H1 or spermidine; lane 5, kDNA of Leishmania donovani and lane 6, kDNA treated with topoisomerase II which releases monomer kDNA minicircles (Chakraborty and Majumder, 1987). (B) kDNA is composed of thousands of mini-circles canenated with maxi-circles (~30 kb) or within mini-circles (~860bp).; (C) The super-coiled, relaxed and linear minicircles were shown that were obtained by digestion of kDNA with topoisomerase II. (D) Plasmid DNA (lane 1) treated with topoisomerase I (lane 2), isolated unique topoisomer from lane 2 was treated with topoisomerase I (lane 3) or topoisomerase II (lane 4) showing linking number changes. All reactions support that DNA has inherent property to make many topological isomers and hence forms nanostructures (Chakraborty et al., 1994).
Watson-Crick ds-DNA has minor groove and major groove and free electron of sugar oxygen and phosphate oxygen providing centres of electrostatic attractions facilitating the formation of nano-particles of different shape and size. The field of structural DNA nanotechnology could be traced back to the words written by Nadrian Seeman in 1982 “It is possible to generate sequences of oligomeric nucleic acids which will preferentially associate to form migrationally immobile junctions, rather than linear duplexes, as they usually do [16] and further advanced by Seeman and co-workers with the construction of relatively flexible branched junction structures and topological structures [17,18] and progressing to the fabrication of crossover DNA tiles with greater rigidity. These DNA tiles could be used to assemble higher-order periodic and aperiodic crystal lattices as the basics of DNA nano-technology [19-23].
One of the most important developments in structural DNA nanotechnology since the introduction of the crossover tile DNA nano-particles was the use of a ‘scaffold’ DNA strand for the assembly of aperiodic structures. It had been previously demonstrated that a long single-stranded DNA chain could be used to organize double-crossover tiles into barcode-patterned lattices, and that a 1.7-kb single-stranded DNA chain could serve as a scaffold for the assembly of a 3-D wire-frame octahedron (Figure 5). The breakthrough came with the concept of ‘DNA origami’, where a long scaffold strand (single-stranded DNA from the M13 phage genome, ~7,429 nucleotides long) was folded with the help of hundreds of short ‘staple’ strands into defined two-dimensional (2-D) shapes [24,25].
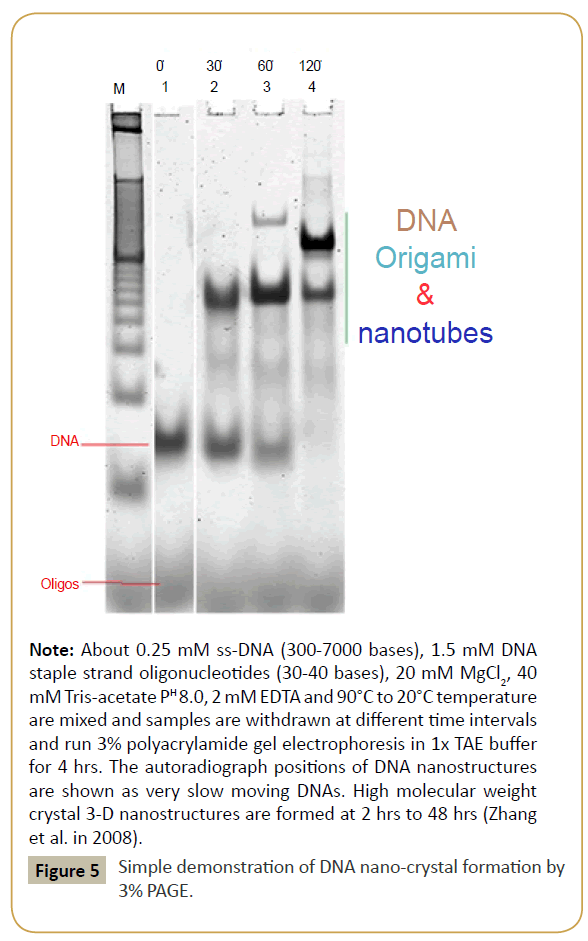
Figure 5: Simple demonstration of DNA nano-crystal formation by 3% PAGE.
Note: About 0.25 mM ss-DNA (300-7000 bases), 1.5 mM DNA staple strand oligonucleotides (30-40 bases), 20 mM MgCl2, 40 mM Tris-acetate PH 8.0, 2 mM EDTA and 90°C to 20°C temperature are mixed and samples are withdrawn at different time intervals and run 3% polyacrylamide gel electrophoresis in 1x TAE buffer for 4 hrs. The autoradiograph positions of DNA nanostructures are shown as very slow moving DNAs. High molecular weight crystal 3-D nanostructures are formed at 2 hrs to 48 hrs (Zhang et al. in 2008).
Preparation of DNA Nanoparticles
Solid DNA is thread like whitish stable structure which is very soluble in water giving viscous opaque solution and could be detected in ng quantities by fluorescence banding of ethidium bromide stained agarose gel or 260 nm UV spectrometry. Large genomic DNA is hard to handle as it breaks into parts due to high viscosity. The starting material for DNA nano-particles was made by synthesising short oligo-nucleotides (two dozen to few dozen phosphate-sugar-base units), called oligonucleotides primer or staple strands which have cross AT/GC pairing with parent single stranded cccDNA strand which is usually M13 viral DNA (~7400bases). Such DNA mixture (1:5 at 1-5 mM concentrations) when heated to 90°C and then slowly cooled 20˚C in presence of ~20 mM MgCl2, 2-D and 3-D ds-DNA nano-structures were formed.
Bacterial genomic DNA could be isolated by digesting bacteria in SDS-Proteinase-K overnight followed by phenol-chloroformisoamyl alcohol (25:24:1) extraction and ethanol precipitation (2 vol., 99% pure) in presence of salt (300 mM NaCl). Then sonicate the genomic DNA to get smaller DNA which could be further purified by agarose gel electrophoresis followed by elution of specified length ds-DNA from gel using DNA Gel extraction kit from Promega or Qiagen. As the genome sequence is known, any DNA staple strands could be synthesized which could form double cross over (DX) or triple cross over (TX) with parent ssDNA giving nano-crystals. Single stranded bacteriophage DNA (M13) was used with success and design of staple strands would be easy and DNA origami of different shapes could be prepared using few to hundred staple strands (30-40 bases; 1-5 mM solution; 10 mM MgCl2) from different part of the M13 genome [26]. Now, different softwares were available which could predict the main DNA (200- 300 bases) and 32 bases long different oligonucleotides straple strands to produce 3-D DNA nano-tubes or DNA nano-cages. One can design many sticky strands like cohesive ends of EcoRI and PstI restriction enzymes digested DNAs or different nob like hairpin structures between AT/GC pairing complementary staple strands which favours the DNA crystal formation at the range 10- 50 nm (Figure 5).
DNA Holiday Junction
Holliday Junction was produced as a DNA recombination intermediate in cells and was proposed by Dr. Rabin Holiday in 1965. Dr. N. C. Seeman in 1980 had shown a number of DNA nanostructures using PX DNA branched staple strands very similar to holiday junction of chromosome recombination (Figure 6). In other words, DNA assembly was the inherent property of DNA itself of B-DNA or Z-DNA and was favoured by Mg++ ions and proteins which could be histones and transcription factors.
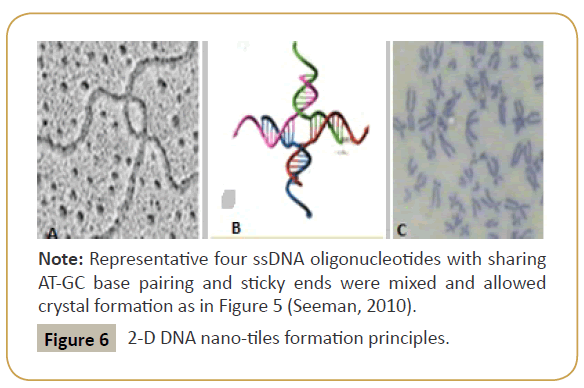
Figure 6: Four staple strands complex and chromosome holiday junction.
Note: Holiday junction formation during chromosomes crossover in cells (panel A) and similar structure was formed in DNA nanotiles (panel B) and original compressed chromosome structure in mammalian cells (Panel C) were presented. In DNA tile four cross sharing oligonucleotides are used shown in different colour. Note that human DNA ~1.5 meter long in 46 chromosomes (3x109 bases) but compacted into 5 micrometer diameter nucleus where each metaphase chromosome DNA appeared as 10-20 nanometer length due to histone assembly, helical structures and electrostatic interactions with non-histone proteins (Chen et al., 2004).
DNA Tiles and DNA Nanotubes
DNA tiles defined as ds-DNA fragments with sticky ends composed from few synthetic oligonucleotides (30-100 bases) with multiple complementary hydrogen bonding motifs among themselves and forms variety programmable DNA lattice structures in presence of 10-20 mM Mg++ ion at 90C to 20C program in a thermostat for 2 hrs [26,27]. Self-assembled 2D DNA tiling lattices composed of tens of thousands of tiles had been demonstrated by Seeman, Winfree, and Reif groups and visualized by molecular imaging techniques such as atomic force microscopy (Figure 7). There are at least three major strategies for the formation of patterned DNA tiling lattice with DX and TX self-assemblies: (i) Unmediated algorithmic self-assembly is the simplest method for 2D pattern formation using 3-10 sets of DNA tiles that self-assemble in a predictable manner. Unmediated algorithmic self-assembly of complex patterns requires delicate control of physical phenomena, which includes nucleation rates, crystal growth rates, spontaneous nucleation, and error rates in solutions containing many distinct DNA tile types but hardly requires patterned oligonucleotides. (ii) 2nd Sequential stepwise assembly of DNA tiles was started with molecular building blocks (MBBs) attached to a solid support, to enable removal of excess reactants after each step. Such sequential assembly might allow for the synthesis of complex molecular structures, while extensively reusing a small number of MBB types. (iii) Directed nucleation assembly is the 3rd method used for selfassembly of complex patterned lattice. It uses a preassembled input DNA strand that encodes the required pattern information; other oligonucleotides then assemble into specified tiles around this input scaffold strand, forming the desired 1D or 2D pattern of DNA tiles. Karina et al. produced Rolling Circle Amplification nanotubes (RCA) using circular phage M13 or phage 29 and 31 base primers (0.4-1.5 mM) led to nanotubes formation in 1x TAE + Mg++ (20 mM) buffer at buffer at 95°->4°C thermostat thermostat [28-31].
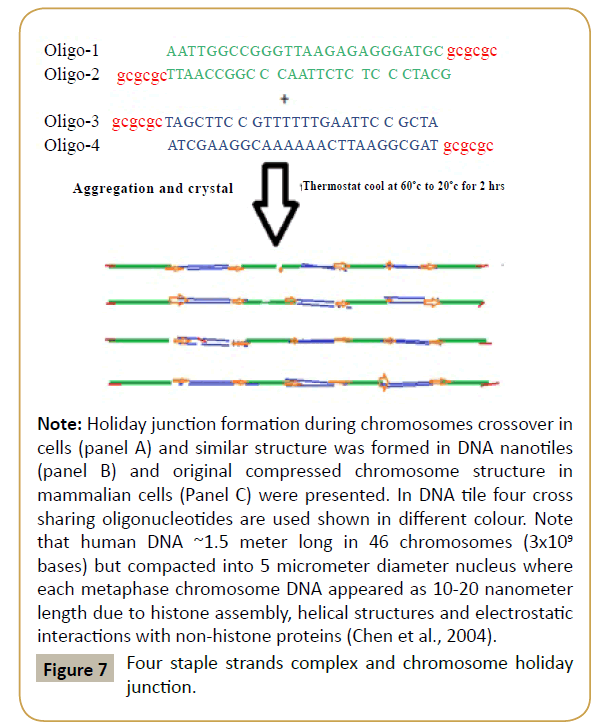
Figure 7: 2-D DNA nano-tiles formation principles.
Note: Representative four ssDNA oligonucleotides with sharing AT-GC base pairing and sticky ends were mixed and allowed crystal formation as in Figure 5 (Seeman, 2010).
DNA Origami
Origami means to a Japanese art of transforming a flat sheet of paper into an arbitrarily shaped object through folding and sculpting techniques. In DNA origami, a long single strand of DNA (scaffold) is folded into various shapes by hundreds of synthetic oligonucleotides, referred to as staple strands targeted to bind to different places along the scaffold anti-parallel DNA strand, giving a precise size and shape of the final DNA nano-structure, called DNA origami structures or origami nanotiles (Figure 8) [32]. As for example 7249 bp M13mp18 DNA with 225 oligonucleotides of 32 nt long could assembled within 2 hrs into 90 nm × 60 nm rectangular origami with 32 helices [19,33,34]. Sugiyama et al. have also reported box structures with similar design features and Yan et al. have used a related strategy to construct a hollow 3-D DNA origami tetrahedron. Shih and co-workers also have reported a new set of design principles for 3D origami construction [34]. Rather than hollow structures, the origami was denser with a design based on the parallel arrangement of helices into a honeycomb lattice. Further, Yan and co-workers have reported a strategy to design and construct 3D DNA origami structures that contain highly curved surfaces providing DNA into under wound or over wound as compared to B-DNA with 10.5 bp per helical turn as well as 3-D tensegrity origami [35-37]. Tensegrity means to construct light weight structures from compressed (rigid) beams connected by stress-bearing wires. When applied to DNA origami, the scaffold is used to generate the rigid beams, together with the staple strands, but also as the stress-bearing wires and is very stable DNA structures [38].
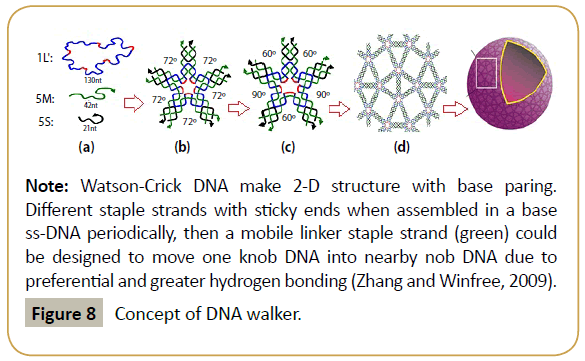
Figure 8: 3-D DNA origami formation principles.
Note: One circular ssDNA (large) and other few staple oligonucleotides are assembled (Zhang et al. 2008; with permission) as in Figure 5 and 3-D nano-tubes are formed (Zheng et al. 2009).
DNA Aptamer
Aptamers are ss-DNA or ss-RNA oligonucleotides that can selectively bind certain targets such as proteins/ nucleic acids/ small organic compounds with high specificity. Selective thrombin DNA aptamer (5’-GGTTGGTGTGGTTGG-3’) binding as tool to position proteins in the self-assembled DNA arrays [38]. Polystyrene adaptamer (PS13-DNA) nanotubes has been characterized at 36 nm when annealed at 95¡C in a thermostat coupling and polymer ultra-structure were confirmed by 3% PAGE and TEM [25,39-42].
DNA Walker
Concept of DNA walker is branched DNA strands located on the parental strand hybrids could be hybridised repeatedly by walking mobile staple strand following strand displacement method and sequestering a move of electrical signal which could be utilized as barcode [43-45]. Green mobile strand could move one hairpin nob into another due to AT-GC pairing and greater strand homology (Figure 9).
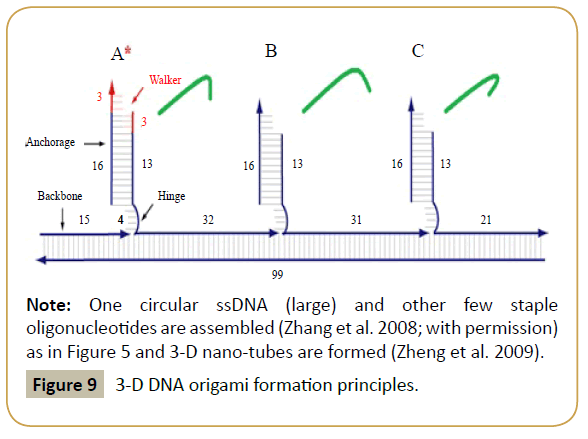
Figure 9: Concept of DNA walker.
Note: Watson-Crick DNA make 2-D structure with base paring. Different staple strands with sticky ends when assembled in a base ss-DNA periodically, then a mobile linker staple strand (green) could be designed to move one knob DNA into nearby nob DNA due to preferential and greater hydrogen bonding (Zhang and Winfree, 2009).
Problems of DNA-Nanoparticles
Working with Ag-nano-particles is quite easy but using DNA, you need a DNAase, RNAase and Protease free environment, and otherwise you will see no result. DNase present in every organism from bacteria to fungus to yeast. So all reagent and chemicals are used in preparing DNA-nanoparticles must be aseptic conditions and must be free from DNase enzyme. Simple way to do it, autoclave at 121C for 15 min or heat at 250C for 2 hrs or wash with ethanol repeatedly (whatever applies to your system) and possibly keep 1-10 mM EDTA in buffer and use gloves during work. So handling with RNA like in ribozyme technology is more serious because RNaseH/T/A also present in every organism and is very stable and active [46]. Similarly protease could cleave proteins used in DNA-nanotechnology. Thus during use of polymer biomolecules like DNA, RNA and protein excess heat and hydrolytic enzymes contaminations must be avoided [47]. Similarly, during each experiment you have to check the preparation by atomic force microscopy to TEM/ SEM as oligonucleotides form crystal structures. However, 3% polyacrylamide gel could be easy to see the slow moving crystals by EtBr staining or autoradiography in case of radiolablled oligonucleotides. Biotin-labelled oligonucleotides could be used using non-radioactive labelling method involving streptavidineconjugated peroxidise.
DNA-Nanotechnology in Medicine
Nanotechnology uses for the treatment, diagnosis and control of diseases has been referred to as “nano-medicine” [48-50]. The work of N. C. Seeman and co-workers have established DNA structures as versatile building blocks for complex nano-scale assembly including immobile holiday junctions, cubic cages, and two dimensional lattices to the vision of a three dimensional DNA crystals. Such DNA-nanoparticles could carry drugs and gene medicines [51]. Although the application of nanotechnology to medicine appears new, the basic nanotechnology approaches for medical application was known for decades. The first example of lipid vesicles or liposomes in drug delivery and DNA transfection were described in 1960s and the first controlled release polymer system of macromolecules was described in 1976 [3] and the first quantum dot bio-conjugate was described in 1998 [5] and the first nanowire nanosenser dates back to 2001 [6]. DNA nanostructures are biocompatible and very potential in drug delivery with no toxicity [52-54]. Carbon monolayer (fullerenes) nanotubes have been increasingly used to deliver plasmids and DNA-conjugates in gene therapy and gene targeting [55].
Uses of DNA nano-particles to cure bacterial infections
The medicinal uses of silver have been documented since 1000 B.C. Silver is a health additive in traditional Chinese and Indian Ayurvedic medicine. Silver nanoparticles broadly accomplish its activities in three ways against Gram-negative bacteria: (1) nanoparticles (~2–8 nm) attach to the surface of the cell membrane disrupting permeability and respiration (2) inside the bacteria they cause further damage by interacting DNA and, (3) nanoparticles release silver ions which could bind cysteine residues. ROS and free radical production is one of the primary mechanisms of nanoparticle toxicity; it may result in oxidative stress, inflammation, and consequent damage to proteins, membranes and DNA for bactericidal activities [56]. Studies demonstrated the potential for nano-materials to cause DNA mutation and induce major structural damage to mitochondria resulting in cell death [57]. The development of drug resistance in bacteria has become a serious problem in public health and alternative methods are welcomed by medical authorities [13]. The structural differences in gram(+) and gram(-) bacteria lie in the organization of the peptidoglycan layer as a thin layer (~2–3 nm) between the cytoplasmic membrane and the outer membrane, in contrast to Gram-positive bacteria lack the outer membrane but have a rigid peptidoglycan layer of about 30 nm thick. A multi-drug resistant strain of gram-negative (Salmonella typhus, resistant to chloramphenicol, amoxicillin and trimethoprim) bacteria was also subjected to analysis to examine the antibacterial effect of the nanoparticles [58]. Silver and gold nanoparticles with Cinnamomum plant extract show very potent antibacterial activities and also cure multi-drug resistant bacterial infections [59,60]. Such advantage when combined with DNA nanotubes, immobilized gold with DNA nanostructures were found more effective in killing pathogens and no toxicity to the host was detected [61,62].
Uses of DNA nano-particles to cure cancer
DNA nanostructures are ideal vehicles to deliver toxic drugs into tumour sites less affecting normal cells. Thus, DNA is genetic material, possesses high biocompatibility and low cytotoxicity, ideal for applications in the biomedical field [63-66]. Its remarkable molecular recognition properties, and GC/AT base pairing with mechanical rigidity, nano-dimensions of the repeating unit, easily custom synthesis with any length of strands allow the formation of any shape of 2-D and 3-D nanostructures with versatile highly nontoxic drug nano-carriers (Figure 10) [67,68]. In 2004, the National Cancer Institute (NCI) in the USA created the Alliance for Nanotechnology in Cancer, which spearheads the integration of nanotechnology into biomedical research through the coordinated effort of a network of investigators from diverse institutions and organizations. The Centres of Cancer Nanotechnology Excellence (CCNEs) integrate discovery and tool development for nanotechnology applications into clinical oncology. CCNEs link physical scientists, engineers, and technologists working at the nanoscale with cancer biologists and oncologists. The Cancer Nanotechnology Platform Partnerships are engaged in directed, product-focused research that aims to translate cutting-edge science and technology into the next generation of diagnostic and therapeutic tools. These platforms serve as core technologies for a wide array of applications. Doxorubicin coupled with isosahedral and hexagonal DNA origami and also with SiRNA used to deliver cancer cells with success [69-72]. Small interfering RNAs of VEGF, XLAP, PGP, MRP-1, BCL-2 and cMyc genes have shown immense promise to knockout drug resistance genes as well as to recover the sensitivity of resistant tumours to anticancer therapy [73-76]. Combinatorial anticancer effects of curcumin and 5-fluorouracil loaded thiolated chitosan nanoparticles towards colon cancer also has been developed [65].
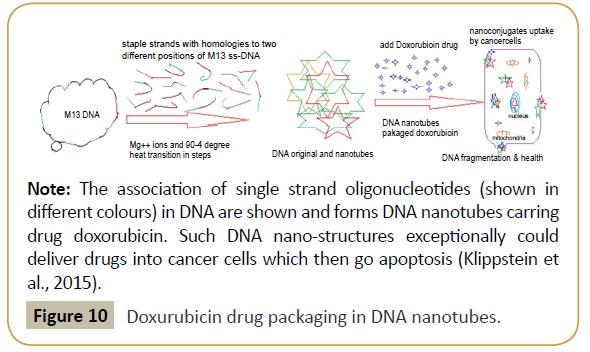
Figure 10: Doxurubicin drug packaging in DNA nanotubes.
Note: The association of single strand oligonucleotides (shown in different colours) in DNA are shown and forms DNA nanotubes carring drug doxorubicin. Such DNA nano-structures exceptionally could deliver drugs into cancer cells which then go apoptosis (Klippstein et al., 2015).
DNA nanostructures used against viral diseases
Versatile nature of DNA nanotubes as carrier of drugs, antisense RNA, enzymes to the target sites also have been utilized to control virus penetration and elimination [77].
Discussion
Pinheiro et al. beautifully have explained in an research article in Nature Nanotechnology in 2011, the advanced nano-chips in modern mobile phone vs. in an ancient calculator and also in a biology context, ~40 proteins and few RNA molecules complexed to form ribosome where protein synthesis occur [9]. Such examples satisfied the future of DNA nanotechnology and to design many advanced technologies with biological particles as well as biophysical particles. The essence is that DNA is also very stable and a good carrier of electrons if protected from heat and nuclease enzyme. Thus, a device like cellulose fibre or like fullerene carbon sheet nanoparticles could be assembled with DNA molecules to increase the stability of DNA nano-particles or DNA nano-transistors [78]. As for example, graphene oxide has been utilized as a veritable wonder material, when incorporated into nano-cellulose foam (as the lab-created bacterial product) conducting heat and electricity quickly and efficiently. The cellulose at the bottom of the bi-layered bio-foam acts as a sponge, drawing water up to the graphene oxide where rapid evaporation occurs and a system for easy water purification was also prepared [59].
Recently, nano-technology has been utilized in many nanomaterials with negative Poisson's ratios with shear resistance and fracture toughness as has been representatively used as vanes for aircraft gas-turbine engines and also applicable to ultrathin oxides films, carbon nano-tube, ferroelectrics, bucklicrystal and fluorides [55]. Boriskina et al. explore different regimes of hybrid optical-thermal antenna operation that depend on the intensity of the light illumination and thermal resistances between the nano-antennas and the rest of the optical nano-chips. The authors also demonstrate how hybrid optical-thermal antennas can be used to achieve strong localized heating of nanoparticles while keeping the rest of the optical chip at low temperature, which can be useful for applications in thermal and thermally assisted catalysis. Plasmonic nanoantennas have opened new horizons in bio-chemical sensing and nanoscale imaging and use of NA nanoparticles could be in the door [79,80]. Recently, Sleiman et al. has integrated a RNA antisense strand of Luciferase in DNA tiles which has reduced the luc gene expression in HeLa cells up to 48 hrs suggesting the use of DNA nano-chips in future molecular biology techniques [81]. DNA nanoparticles mediated vaccine delivery could be a good promise [82].
Conclusion
DNA nanotechnology has given good promise in drug delivery and preparation of biochips and biosensors. However, heat and nuclease sensitivities are much problem and many conjugated adaptamer methods have been formulated like locked nucleotides, peptide nucleic acids, L-DNA, DNA-RNA hybrids, PEG-nucleic acids, Cellulose/Cholesterol/polystyrene-adaptamer oligonucleotides and so many more. UNIQUIMER 3D software has been tested on the design of both existing motifs (holiday junction, 4 × 4 tile, double crossover, DNA tetrahedron, DNA cube, etc.) and non-existing motifs (soccer ball) and other complex DNA nanostructures [72]. We foresee a challenging golden era of DNA nanotechnology in medicine and biology. Use of DNA nanostructures in DNA Walker, DNA Chips, DNA Robotics, DNA Transistors are other applications where we will see tremendous success in the future [52,53].
Acknowledgement
We thank Dr. Bidyut Bandhopadhyay for help during the study and Dr. J. B. Medda for financial support.