Research Article - (2016) Volume 2, Issue 4
Masahiko Tanabe1,3*, Sally Fujiyama2 and Yoshiya Horimoto3
1Department of Breast Surgical Oncology, Kyoundo Hospital 1-8, Kandasurugadai, Chiyoda-ku, Tokyo, 101-0062, Japan
2Department of Applied Biological Science, Graduate School of Agriculture, Tokyo University of Agriculture and Technology 3-5-8, Saiwai-cho, Fuchu-shi, Tokyo, 183-8509, Japan
3Department of Breast Oncology, Juntendo University School of Medicine 2-1-1, Hongo, Bunkyo-ku, Tokyo, 113- 0033, Japan
*Corresponding Author:
Tanabe M
Department of Breast Surgical Oncology, Kyoundo Hospital 1-8
Kandasurugadai, Chiyoda-ku, Tokyo, 101-0062, Japan
Tel: +81-3-3292-2051
Fax: +81-3-3292-3376
E-mail: mtanabe-tky@umin.ac.jp
Received date: October 18, 2016; Accepted date: October 26, 2016; Published date: October 31, 2016
Citation: Tanabe M, Fujiyama S, Horimoto Y. Developmentally regulated GTP binding protein 2 (DRG2) and Nup107 are associated with epigenetic regulation via H2A.Z on promoter regions of specific genes in MCF7 breast cancer cells. J Clin Epigenet. 2016, 2:4. doi: 10.21767/2472-1158.100029
Copyright: © 2016 Tanabe M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Background: Epigenetic changes are critical events in breast cancer development. The regulation of epigenetic marks is orchestrated by DNA methylation, modifications of canonical histones, and incorporation of histone variants. A histone variant of H2A.Z was recently reported to be associated with breast cancer progression. However, the physiological function of H2A.Z in breast cancer has yet to be revealed. We previously identified genetic interactions between H2AvD (Drosophila H2A.Z) and DRG2 by means of Drosophila screening.
Results: We herein focused on identifying new factors associated with DRG2 in order to reveal the epigenetic impact of H2A.Z on breast cancer. Initially, in a series of protein purification assays with a Drosophila cell line, we identified Nup107, a nuclear pore complex protein, as one of the factors interacting with DRG2. In addition, RWD domain containing 1 (RWDD1), GCN1 like protein 1 (GCN1L1), eukaryotic translation initiation factor 4B (eIF4B), and argininosuccinate lyase (ASL) were co-purified. Next, employing chromatin immunoprecipitationmicroarray analysis on the human breast cancer cell line MCF7, we revealed that H2A.Z, DRG2, and Nup107 co-occupy the promoter regions of 1,947 genes. In the common genes showing co-occupation with these three factors, the largest category, which included 871 genes, was found to consist of ‘alternative splicing variants’. This result and the factors co-purified suggest that H2A.Z, DRG2, and Nup107 co-regulate mRNA splicing as well as nuclear export and translation.
Conclusions: Further analysis focusing on the biological functions of DRG2 and Nup107 would facilitate elucidating the epigenetic impact of H2A.Z on breast cancer development.
Keywords
Breast cancer; H2A.Z; Estrogen; Estrogen receptor; DRG2; Nup107; Epigenetics
Abbreviations
ASL: Argininosuccinate Lyase; ChIP: Chromatin Immunoprecipitation; ChIP-chip: ChIP-Microarray Analysis; DRG2: Developmentally Regulated GTP Binding Protein 2; eIF4B: Eukaryotic Translation Initiation Factor 4B; E2: Estrogen; ER: Estrogen receptor; GCN1L1: GCN1 like protein 1; RWDD1: RWD Domain containing 1; S2 cells: Drosophila Schneider cells
Background
Breast cancer is caused by the accumulation of somatic mutations, but factors that directly regulate disease progression are as yet poorly understood [1]. It is well known that approximately 90% of breast cancers are sporadic. Therefore, it is speculated that epigenetic changes without sequential mutation of DNA, along with somatic mutations, are themselves significant. Epidemiological studies have demonstrated that environmental, dietary, and lifestyle factors impact the occurrence of breast cancer, and it is well known that high lifetime exposure to estrogen (E2) increases the risk of breast cancer [2]. E2 plays critical roles in the initiation and development of breast cancer [3,4]. Biological functions of E2 are mediated through estrogen receptor (ER) α/β. ERs are ligand-dependent transcriptional factors essential for the expressions of ER-target genes. Epigenetic changes of ERα-target genes are critical for the development and progression of breast cancer [3]. However, the mechanism by which a single epidemiologic factor, such as E2 stimulation, can alter epigenetic marks on ER target genes has not been elucidated. We are interested in alterations of epigenetic marks, in response to lifetime E2 exposure, which may potentiate breast cancer initiation and development.
In addition to DNA methylation and modifications of canonical histones, several histone variants function as epigenetic marks in gene expression [5-9], meaning that regulation of gene expressions includes the replacement of canonical histones with histone variants as well as post-translational modifications of the tails of both the canonical histones and their variants [5]. H2A.Z, one such histone H2A variant, was reported to be essential for transcriptional activation of ERα-target genes [10]. Recently, the clinical impacts of H2A.Z on breast cancer development have attracted attention [1,5,11]. High expression of H2A.Z was shown to be significantly associated with breast cancer proliferation, development [11], progression and lymph node metastasis [12]. However, the molecular mechanism by which H2A.Z regulates the proliferation and progression of breast cancer has yet to be revealed.
H2A.Z, which is highly conserved from yeast to humans, was experimentally confirmed to be essential for survival in Drosophila and mice [13,14], suggesting that H2A.Z has important functions that cannot be carried out by canonical histones [5]. H2A.Z is mainly localized in active gene promoters and regulates gene expressions [6-9]. While the Drosophila H2A.Z homologue of H2AvD has been well studied in terms of its function and interactions with other factors, by both genetic and biochemical approaches, there has been little investigation of the biological functions of human H2A.Z. Thus, we employed several Drosophila screening systems and protein purification methods, as previously described [15-20], in efforts to identify novel factors associated with epigenetic regulation that is dependent on H2AvD. Employing these experimental approaches, we previously identified developmentally regulated GTP binding protein 2 (DRG2) as a novel factor genetically associated with H2AvD. The DRG2 gene encodes a 364 amino acid protein involved in cell proliferation and differentiation [21-24]. However, to our knowledge, there have been no studies demonstrating DRG2 to function in the nucleus. Therefore, whether DRG2 associates with H2A.Z/H2AvD on chromatin remains unknown. Herein, we investigated the molecular function of DRG2 in the nucleus, and revealed the mechanism by which DRG2 associates with H2A.Z by applying protein complex purification and chromatin immunoprecipitation-microarray analysis.
Materials and Methods
Purification of protein complex: Biochemical protein complex purification was performed as described in previous reports [15-18]. Drosophila Schneider (S2) cells were cultured in Schneider’s Drosophila Medium (Gibco BRL, Grand Island, NY) supplemented with 10 % fetal bovine serum at 25 ?. For purification of the Drosophila DRG2 (dDRG2)-associated complex, S2 cells stably expressing FLAG-His-dDRG2 were established by co-transfection of FLAG-His-tagged-dDRG2 expression plasmid cloned into the pAct vector (Promega, Madison, WI; GenBank accession no. AF264723) and pCoBlast vector (Invitrogen), and were selected by adding 25 mg/mL Blasticidin. Nuclear extracts of S2 cells (Mock) and FLAG-His-dDRG2 expressing S2 cells, which had been prepared as described previously [15-18], were incubated at 4? with 40 μL of anti-FLAG M2 agarose (Sigma) for 6 h. After being washed five times with buffer D (20 mM HEPES-KOH, 0.2 mM EDTA, 5 mM MgCl2, 150 mM KCl, 0.05% [v/v] NP-40, 10% [v/v] glycerol, 0.5 mM DTT, 0.2 mM PMSF, protease inhibitor cocktail [Roche] at pH 7.9), the protein complex was eluted with 200 μg/ ml FLAG peptide (Sigma) in buffer D. Ni-NTA Agarose (QIAGEN) was used for further His-tag affinity purification. The samples obtained were analyzed by SDS-PAGE followed by silver staining.
Glycerol density gradient centrifugation: In order to fractionate protein complexes according to their molecular weights, glycerol density gradient centrifugation was performed as previously described [15]. The eluted complex was fractionated by 10–50% glycerol density gradient centrifugation at 4?, 182,000 g using a SW-40 rotor (Beckman Coulter) for 16 h. Gradients were fractionated into 1 mL each and the obtained fractions were analyzed by Western blotting with the antibody against DRG2 (14743-1-AP, Proteintech). The fractions containing the dDRG2 complex were analyzed with SDS-PAGE followed by silver staining.
Protein identification: The silver stained bands derived from factors interacting specifically with dDRG2 were subjected to ingel digestion and peptide mass fingerprint analysis using mass spectrometry. Sliced gel-bands containing proteins were incubated in 10 mM DTT and 100 mM NH4HCO3 at 56 ? for 1 h, and cysteine SH groups were alkylated with 55 mM iodoacetamide and 100 mM NH4HCO3 at room temperature for 45 min. After alkylation, the gels were washed with acetonitrile and 100 mM NH4HCO3 alternately and incubated with trypsin gold (Promega) for protein digestion overnight at 37?. The peptides were extracted from the gels with 5% formic acid and 50% acetonitrile. The resulting peptide mixtures were then applied to disposable pre-spotted Anchor Chip MALDI target (Bruker Daltonics) and analyzed using Ultraflex MALDI-TOF/TOF mass spectrometry (Bruker Daltonics).
Chromatin immunoprecipitation (ChIP) and ChIP-microarray analysis (ChIP-chip): ChIP analysis using a breast cancer cell line of MCF7 was performed according to the protocol previously reported by ACTGen (Nagano, Japan) [25]. Antibodies of DRG2 (sc164233, Santa Cruz biotechnology), Nup107 (ab85916, Abcam), and H2A.Z (ab4174, Abcam) were used for ChIP analysis. ChIP-chip service and analysis were performed at MBL (Nagoya, Japan). The Roche Nimblegen Human ChIP-chip 3x720K RefSeq Promoter array was used.
Results
Purification of a dDRG2-associated complex and identification of complex components
The DRG2 gene is highly conserved from Drosophila to humans and Drosophila DRG2 (dDRG2) protein is almost identical to human DRG2 protein [21].
To explore the novel factors interacting with dDRG2, which might shed light on its molecular functions in the nucleus, we endeavored to biochemically purify a dDRG2-associated complex. As the material for affinity purification, a Drosophila S2 cell line stably expressing the FLAG-His-tagged-dDRG2 protein was established. First, the nuclear extract, which had been extracted from the nucleus with 210 mM NaCl, was placed in contact with anti-FLAG M2 antibody agarose. After washing the agarose, purified factors were eluted with FLAG peptides. The derived eluent was subjected to further purification with nickel affinity resin for His-tag affinity purification (Figure 1(a)). The eluted sample was subjected to SDS-PAGE and silver staining (Figure 1(b)). Next, we applied more severe conditions to achieve nuclear extraction with 420 mM NaCl. The nuclear extract was placed in contact with anti-FLAG affinity agarose, and the eluted sample was subjected to SDSPAGE and silver staining (Figure 2(a),(b)). Several specific bands were observed in the samples derived from dDRG2-expressing cells, but not in those from the mock-purified samples from wild type S2 cells (Figures 1(b) and 2(b)). The gel bands specific to the dDRG2 lane were excised and then subjected to identification by in-gel digestion followed by the peptide mass fingerprint method using MALDI-TOF/TOF mass spectrometry. This assay identified seven proteins, i.e. CG17514 / GCN1L1 (Figure 1(b)), heat shock protein cognate 4, CG9510 / ASL, isoforms B and E of eIF4B, RWDD1 (Drosophila RWDD1 (dRWDD1), and CG31974 (Figure 2(b)). RWDD1 has already been reported as a binding partner of DRG2 [24], indicating our complex purification and protein identification experiments to have yielded reliable results. Furthermore, we attempted to isolate and identify the protein complex including dDRG2 protein. The extract purified with anti-FLAG antibody immunoprecipitation was fractionated by means of glycerol density gradient centrifugation (Figure 3(a)). The obtained samples were subjected to Western blotting with anti-DRG2 antibody and silver staining (Figure 3(b)). The dDRG2 protein was detected in fractions of 2-8 as well as 11-12, suggesting that dDRG2 forms multi-subunit complexes. Considering that the lowest fraction (11 and 12) represents products of protein aggregation, we focused on fraction 5 and endeavored to identify its components by applying mass spectrometry. This assay newly identified Nup107 (Drosophila Nup107 (dNup107)), in addition to dRWDD1, and the bait protein dDRG2 (Figure 3(c)). Nup107 is a component of the nuclear pore complex and is a member of the nucleoporin family [26]. dDRG2, dRWDD1, and dNup107 were fractionated into the same fraction on the glycerol density gradient, suggesting that these factors existed as one protein complex.
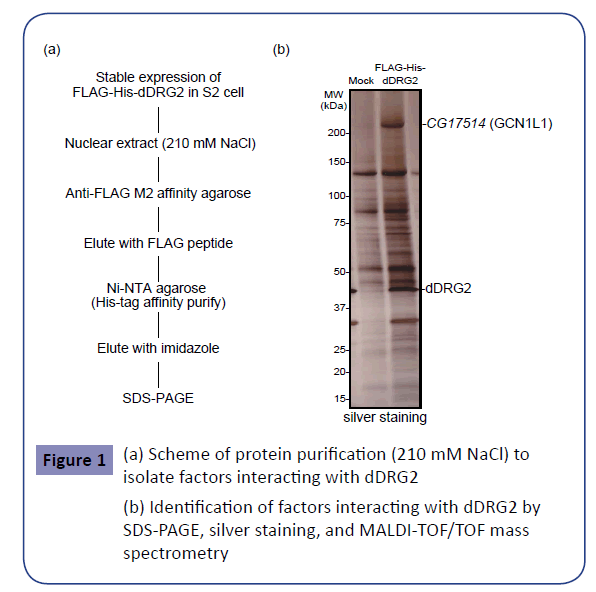
Figure 1: (a) Scheme of protein purification (210 mM NaCl) to isolate factors interacting with dDRG2
(b) Identification of factors interacting with dDRG2 by SDS-PAGE, silver staining, and MALDI-TOF/TOF mass spectrometry
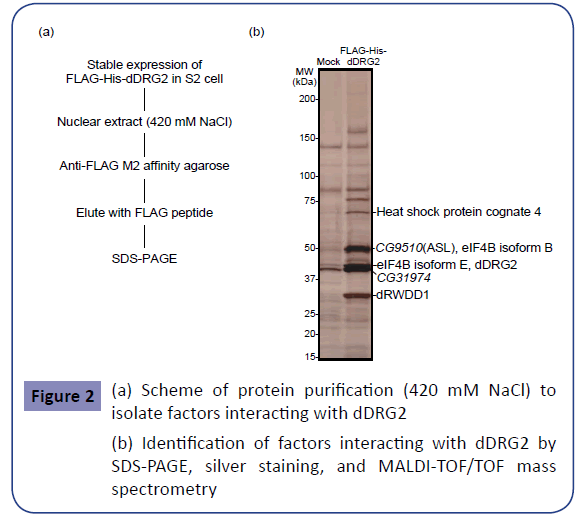
Figure 2: (a) Scheme of protein purification (420 mM NaCl) to isolate factors interacting with dDRG2
(b) Identification of factors interacting with dDRG2 by SDS-PAGE, silver staining, and MALDI-TOF/TOF mass spectrometry
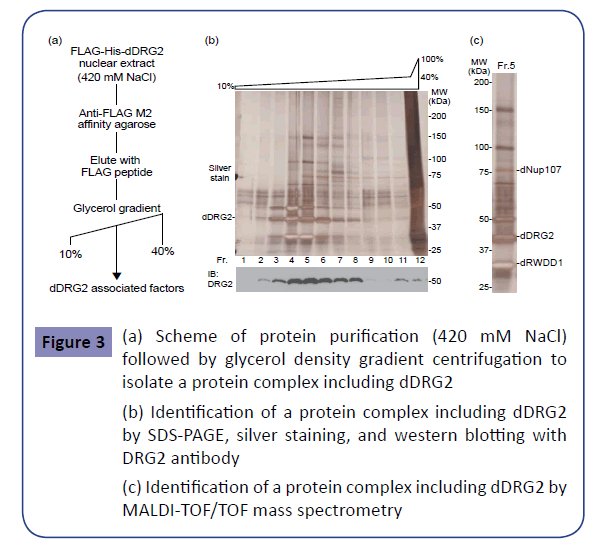
Figure 3: (a) Scheme of protein purification (420 mM NaCl) followed by glycerol density gradient centrifugation to isolate a protein complex including dDRG2
(b) Identification of a protein complex including dDRG2 by SDS-PAGE, silver staining, and western blotting with DRG2 antibody
(c) Identification of a protein complex including dDRG2 by MALDI-TOF/TOF mass spectrometry
H2A.Z, DRG2, and Nup107 co-localized in the promoter region
To address whether these identified factors function together with H2A.Z in the same promoter regions of target genes, we performed ChIP analysis using the specific antibodies for H2A.Z, DRG2, and Nup107 in the human breast cancer cell line MCF7. The samples obtained by ChIP analysis were subjected to ChIP-chip analysis on the Roche Nimblegen Human ChIP-chip 3x720K RefSeq Promoter array. This assay revealed H2A.Z, DRG2, and Nup107 to be located on promoter regions of 8534, 9146 and 8551 genes, respectively (Table 1(a); upper). As shown in the Venn diagram, there were 1947 gene promoters immunoprecipitated by all three antibodies, i.e., those against H2A.Z, DRG2, and Nup107 (Table 1(a)). We next analyzed the functional characteristics of these 1947 common genes by using DAVID bioinformatics resources. The largest category, which included 871 genes, was comprised of ‘alternative splicing variants’ defined as genes which have at least two isoforms due to pre-mRNA splicing. The secondlargest category, which included 809 genes, was comprised of ‘phosphoproteins’ defined as genes coding proteins that are post-translationally phosphorylated. Other characteristics of the identified genes are shown in Table 1(b).
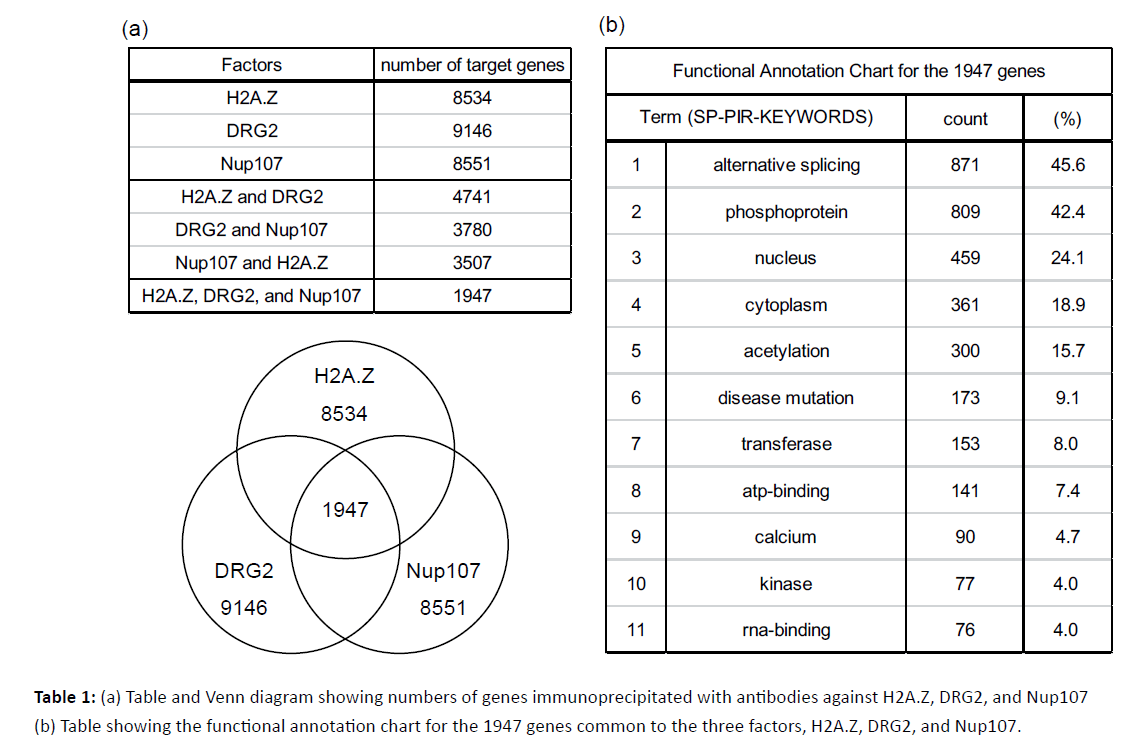
Table 1: (a) Table and Venn diagram showing numbers of genes immunoprecipitated with antibodies against H2A.Z, DRG2, and Nup107 (b) Table showing the functional annotation chart for the 1947 genes common to the three factors, H2A.Z, DRG2, and Nup107.
Discussion
The possibility of direct interaction between H2A.Z and DRG2
In this study, we identified Nup107 as a factor physically interacting with DRG2, which interacted genetically with H2A.Z. In addition, we identified several factors supporting our speculations as to the functions of these three factors.
Although our results revealed genetic interactions between H2AvD and dDRG2 in Drosophila, we detected no physical interaction between H2A.Z and DRG2 in MCF7 cells by immunoprecipitation assay (data not shown). In fact, to our knowledge, there have been no reports focusing on direct interactions between H2A.Z and DRG2, though DRG1 protein reportedly interacts directly with the nucleosome including H2A.Z based on the results of nucleosome pull-down and proteomic analysis [27]. DRG1 protein is similar to DRG2 protein, and 58% of amino acid sequences in human DRG1 and DRG2 are reportedly identical [21]. Based on these observations, we can reasonably speculate that nucleosomes including H2A.Z have the potential to interact with DRG2.
Possible functions of DRG2 and RWDD1 in regulating epigenetics
DRG2 protein has a domain consisting of characteristic G-motifs that may serve as the core of GTPase activity, along with the C terminal TGS domain which is related to RNA binding [21]. Therefore, we speculate that DRG2 associates with mRNA and thereby plays roles in transcription, mRNA splicing, and translation. RWDD1, which is known to interact with DRG2, was reported to interact with androgen receptors and to act as a coactivator of androgen-dependent transcription [28-30]. If RWDD1 can function as a coactivator of other nuclear receptors including ERα/β, it would raise the possibility that DRG2 and RWDD1 function cooperatively in the transcription of ERα target genes.
Nup107 is a component of the nuclear pore complex (NPC). Nup107 has a leucine zipper, which may interact directly with DNA [26]. NPCs localize at the nuclear membrane and mainly function to regulate nuclear transport between the nucleoplasm and the cytoplasm. In addition, NPCs also function in gene activation and chromatin organization [31-33]. Therefore, there is a possibility that DRG2 and Nup107 are cooperatively associated with H2A.Z in nuclear export as well as transcription. As an example, it has been reported that, in yeast, associations between H2A.Z and NPCs regulate transcriptional memory, allowing early reactivation of transiently repressed genes anchored around the nuclear periphery [34].
The other proteins identified, GCN1L1 and eIF4B, also support our hypothesis that the complex functions as a mediator of transcription as well as facilitator of translation [35-38]. We recognize a very general consistency between the functions of the proteins identified and the characteristics of target genes regulated by H2A.Z, DRG2, and Nup107, as shown in Table 1(b).
The possible function of H2A.Z in cancer
Taking our results together with those of previous reports on H2A.Z, we speculate that the physiological function of H2A.Z in breast cancer might be associated with not only modulation of transcription but also mRNA splicing, nuclear export, and translation (Figure 4). Recently, epigenetic changes including H2A.Z have attracted attention as potential treatment targets [1,39-42]. Because altered expression patterns of H2A.Z as well as other H2A variants are associated with the development of several cancers, further analyses focusing, for example, on the implementation of high resolution mass spectrometry for each H2A variant [5], should be conducted in an effort to identify new therapeutic targets for breast cancer as well as other malignant tumors.
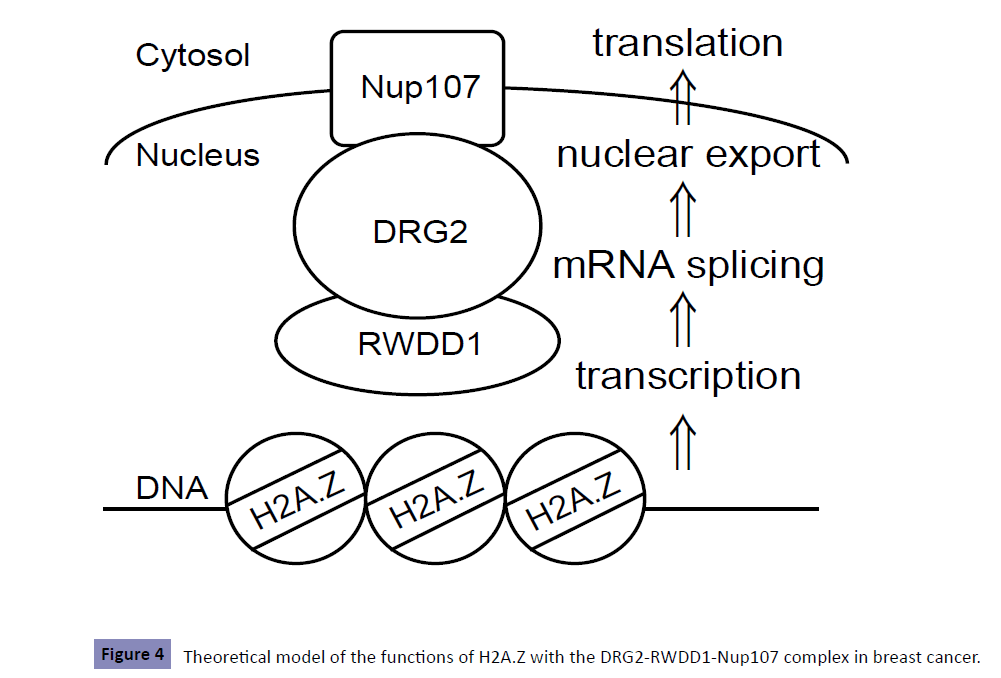
Figure 4: Theoretical model of the functions of H2A.Z with the DRG2-RWDD1-Nup107 complex in breast cancer.
Conclusions
We revealed that DRG2 and Nup107 appear to be associated with epigenetic regulation via H2A.Z involving the promoter regions of specific genes in MCF7 breast cancer cells. DRG2 and Nup107 might be promising factors for elucidating the epigenetic impact of H2A.Z on breast cancer progression based on epigenetic alterations regulated by epidemiologic factors, such as lifetime exposure to E2 stimulation.
Declarations
Ethics approval and consent to participate, Ethics approval and consent were unnecessary for the use of samples in this research.
Competing interests
The authors have no competing interests to declare.
Funding
This work was supported by JSPS KAKENHI (Grant-in-Aid for Young Scientists (B)); Grant Number JP 22770002. The authors declare that there was no other financial support for this research.
Authors’ contributions
MT wrote the manuscript. MT, SF, and YH edited the manuscript. MT and SF designed and/or performed the experiments.
Acknowledgements
We express our gratitude to Shun Sawatsubashi, Takuya Murata, Shuhei Kimura, and Saya Ito for their generous instructions and help with Drosophila genetics and protein purification. We also thank Yuki Imai, Kaoru Mogushi, Misa Imai, and Kousuke Ishikawa for helpful discussions pertaining to this research.