Keywords
Pharmacogenetic; Breast cancers; Antioestrogenic
Introduction
Tamoxifen, a selective oestrogen receptor modulator, has been the mainstay for the therapy and prevention of recurrence in pre-menopausal women with oestrogen receptor-positive breast cancers for almost 40 years. Tamoxifen decreases the risk of relapse by half and the mortality rate by nearly a third in patients with ER (+) breast cancer which count for approximately 80% of all breast cancers [ 1-3]. While aromatase inhibitors are more effective for treating post-menopausal patients with ER (+) breast cancer, tamoxifen was approved by the FDA for metastatic breast cancer and as an alternative to aromatase inhibitors for the chemoprevention in post-menopausal women [4-6]. Hence, tamoxifen remains the gold standard for ER (+) breast cancer. However, tamoxifen efficacy varies widely, many breast cancer patients with adjuvant tamoxifen remain free of recurrence but 30-50% suffer relapse [7-10].
Literature Review
Metabolism of tamoxifen
The pro-drug, tamoxifen, is extensively metabolised by CYP2D6 into therapeutically-active moieties, 4-hydroxytamoxifen and endoxifen; their affinity for the oestrogenreceptors is ~100-fold greater than tamoxifen and their antioestrogenic potency in suppressing ER-dependent cell proliferation is 30-100-fold stronger than tamoxifen [ 6,9,11-15].
Although factors underlying the ineffectiveness of tamoxifen are yet to be fully defined, these active metabolites play a key role in the high degree of interindividual differences in response to tamoxifen therapy [7-10].
Tamoxifen is metabolised via two routes; 4-hydroxylation and N-demethylation which account for ~7% and ~92% of tamoxifen metabolism, respectively. The 4-hydroxylation pathway, which is catalysed primarily via CYP2D6, leads immediately to the formation of 4-hydroxy-tamoxifen. Ndemethylation results in the formation of N-desmethyltamoxifen and is catalysed mainly via CYP3A4/5, and is followed by CYP2D6-mediated secondary metabolism to endoxifen (Figure 1). However, endoxifen can be formed from both N-desmethyl-tamoxifen and 4-hydroxy-tamoxifen and it has been found to play a more significant antiproliferative role than 4-hydroxy-tamoxifen in breast cancer [13,16-19]. Thus, it has been hypothesised that single nucleotide polymorphisms (SNPs) in the highly-polymorphic CYP2D6 can affect tamoxifen biotransformation and therefore may have the potential to predict the clinical outcome of tamoxifen therapy [9,19,20].
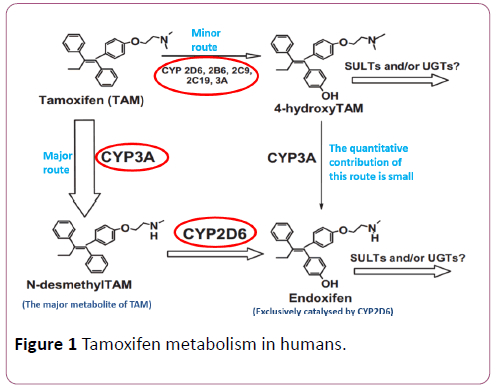
Figure 1: Tamoxifen metabolism in humans.
Cyp2d6 genotypes and predicted phenotypes
More than 100 variants, including more than 20 poor metaboliser alleles, with significant interethnic differences were identified [16,21,22] (Table 1).
| CYP2D6 variant |
Predicted enzymatic function |
Caucasian (Europe) |
Caucasian (USA) |
African-American |
| *1 |
Normal |
33-36% |
27-40% |
29-35% |
| *2 (35%) |
Normal |
22-33% |
26-34% |
18-27% |
| *3 |
Deficient |
1-4% |
1-1.4% |
<1% |
| *4 |
Deficient |
12-23% |
18-23% |
6-9% |
| *5 |
Deficient |
2-7% |
2-4% |
6-7% |
| *6 |
Deficient |
1-1.4% |
1% |
<1% |
| *9 |
Decreased activity |
0-2.6% |
2-3% |
<1% |
| *10 |
Decreased activity |
1.4-2% |
2-8% |
34% |
| *17 |
Decreased activity |
|
|
15-26% |
| *41 |
Decreased activity |
20% |
- |
- |
Note: The most clinically significant variants are circled in red
Table 1: Allelic frequencies of CYP2D6 variant in selected population.
CYP2D6 different genotypes have been associated with four predicted phenotypes based on their CYP2D6 enzymatic function: poor (PM), intermediate (IM), extensive (EM) and ultra-rapid (UM) metabolizers (Table 2).
| Metabolizer status |
CYP2D6-derived genotypes |
Ilepacted effect |
| Ultra-rapid Metabolizer (UM) |
Carrying more than two copies of normal functional alleles |
Higher than expected concentrations of Tamoxifen metabolites at usual doses, possibly adverse reactions |
| Extensive metabolizers (EM) |
Carrying two copies of "normal" function alleles |
Normal |
| Intermediate metabolizers (IM) |
Carrying two reduced function alleles or one functional wild-type and one reduced function allele |
Response between those of EMs and PMs |
| Poor metabolizers (PM) |
Carrying only no-or low-function alleles/Homozygous or compound heterozygous for null alleles |
Possibly non- response and increased risk for breast cancer recurrence |
Table 2: CYP2D6 enzymatic function.
Evidence
The widely-accepted criteria used to evaluate pharmacogenetic tests are: analytical validity, clinical validity and clinical utility [23].
Analytic validity
The heterogeneity at the CYP2D6 locus and the high homology of CYP2D6 locus with its pseudogenes render analyzing CYP2D6 using the widely available short-read “nextgeneration” sequencing platforms to identify duplicated alleles immensely challenging. Nevertheless, clinical “nextgeneration” sequencing and genotyping pipelines have been developed and validated for CYP2D6 pharmacogenetic testing. For example, microarray technology-based genotyping for common CYP2D6 alleles can be optimal with sensitivity and specificity up to 100% [6,24].
Clinical validity
While comparative double-blinded trials showed no correlation between tamoxifen plasma concentrations and clinical response in women treated with tamoxifen [25], plasma levels of endoxifen in those with reduced CYP2D6 activity were found to be considerably low in comparison with those whose CYP2D6 activity is normal [6,16,17,26]. However, since tamoxifen efficacy is dependent upon multiple factors, one of which may be endoxifen plasma level, the variability in clinical outcome of tamoxifen therapy cannot always be explained by CYP2D6 genotype. Furthermore, the precise clinical role of endoxifen in the overall efficacy of tamoxifen is debatable. Endoxifen plasma concentrations cannot be regarded as a surrogate outcome for clinical response and the association between endoxifen concentrations and breast cancer outcomes should be confirmed [27]. Some drug models suggest that both tamoxifen and endoxifen overwhelm oestrogen receptors and therefore small concentrations of endoxifen may be needed to block oestrogen receptors suggesting that variations in endoxifen plasma concentrations may not affect this mechanism [28]. However, endoxifen and other tamoxifen metabolites were proposed to have different mechanisms in modulating hormone receptors [29]. The interpatient variability of tamoxifen metabolism and clinical outcomes in breast cancer may also result from genetic variation in enzymes involved in tamoxifen metabolism such as CYP3A, or resistance to chronic tamoxifen treatment due to activation of somatic mutations in the estrogenic receptor (ESR1) [2,30,31].
Various studies regarding treatment outcomes in the adjuvant and/or metastatic settings have reported that those with genetically impaired CYP2D6 appear to have experienced significantly higher rate of breast cancer recurrence and mortality than those with normal CYP2D6 enzymatic activity [ 10,20,32-38], suggesting that variants associated with reduced function alleles (e.g.*4/*10/*41) are robust predictors of tamoxifen’s antitumoral efficacy [10,20,34,37]. Moreover, findings from a prospective study found that CYP2D6 polymorphisms were strongly associated with Ki-67 response, implying that CYP2D6 genetic variation is an important predictor for efficacy of tamoxifen in women with breast cancer [38]. However, Ki-67 response has not yet been validated as a surrogate marker for tamoxifen efficacy.
Most of these studies were retrospective [33,34,39] and therefore liable to substandard documentation. The prospective studies [20,32], however, were also retrospective in nature since CYP2D6 genotyping has been performed after the relapse had eventuated. Nonparametric analyses and substantial overlaps in the patients’ data were also reported in the following studies [10,33-35]. Nevertheless, two prospective yet small studies [36,38] and two large trials (n>1300) [35,40] have reported statistically strong correlation between CYP2D6 genotype and clinical outcome in patients with breast cancer taking tamoxifen. Although some studies assessed overall survival outcomes [34,39], the trials were mostly non-randomized and failed to show a statistically significant correlation between overall survival and CYP2D6 genotype and whether this association varies by therapy type. In the metastatic setting, however, a significant association was observed but the study was small and the evidence was equivocal and limited [20]. The small sample size and the low coverage of alleles genotyped in most of these studies have diminished the reliability of the tests and reduced the ability to identify false positive results. The insufficient allelic coverage can cause patient misclassification. However, it can be argued that a clear bias toward positive results was observed with higher allelic coverage [41]. Furthermore, the inclusion of ER/PR-negative patients, who do not response to tamoxifen, might have resulted in underestimation of the impact of genotype on clinical outcome and potentially false negative results in some studies [28,41]. These studies have also assumed that CYP2D6 genotype has no impact on clinical outcomes in women with breast cancer who are not taking tamoxifen. The apparent disparity in the ethnic composition, sample size, study design, methodologies, therapeutic regimens, adherence to treatment, co-treatments with CYP2D6 inhibitors and the inconsistent inclusion criteria of the majority of these trials can partly explain the controversial results and render comparison difficult. This has been further complicated by the inconsistent genotype-phenotype assignment and extensive heterogeneity in definition of outcome, as many measured the recurrence or disease-free survival but only a very few assessed the overall survival. More importantly, none of these studies has suggested detailed therapeutic paradigms, or addressed the potential consequences of CYP2D6 testing-based guidance on individuals with phenotypes determined incorrectly.
In contrast, two studies (n=226 and n=677) reported conflicting findings and demonstrated significantly reverse association between clinical outcome of tamoxifen and low activity CYP2D6 genotype [42,43]. It can, however, be argued that using a high dose of tamoxifen in these trials [42,43] might have normalized the low CYP2D6 activity in PMs and IMs. Analyses from two of the most influential large prospective randomized studies (n=1243 and n=588), BIG 1-98 and ATAC, found a near-null correlation between CYP2D6 genotype and breast cancer prognosis in tamoxifen-treated women, and provided statistically strong evidence of the opposite [7,44]. However, this null association was also unexplainably observed in the patients receiving tamoxifen in combination with other anti-tumour agents [45]. It has been suggested that the DNA extracted from formalin-fixed paraffinembedded tumour tissues used in these studies instead of germline DNA [7,44] has potentially led to erroneous genotyping due to the presence of loss of heterozygosity (LOH) at the unstable CYP2D6 locus in breast tumour DNA. This bias was detected by the massive deviation from Hardy-Weinberg equilibrium (i.e. expected versus observed allelic frequencies) [ 46-49]. However, departure from Hardy-Weinberg equilibrium has not been reported in several trials using DNA derived from breast tumour–infiltrated tissues [50]. Even though germline DNA was used or blood samples with (LOH) were excluded, no positive association between genetic variations of CYP2D6 and tamoxifen efficacy was observed [51-53]. Therefore, results from the abovementioned studies [7,44] can be considered valid and should be included in evaluating the value of CYP2D6 genotyping in tamoxifen therapy [38,50,54-56].
To date, there are no standards to validate quality of pharmacogenetic association. “Validation of a true pharmacogenetic association, demonstration of so-called ‘clinical validity’, should be conducted in prospective clinical studies that are larger, have more homogeneous patients and treatment and systematically collect outcomes data” [31].
Clinical utility
Even though it has been recognized that there are certain circumstances in which CYP2D6 genotyping can be helpful for tamoxifen dose adjustment or consideration of alternative treatment [57], CYP2D6 testing is not currently recommended for women for whom tamoxifen treatment is being considered.
Several studies provided evidence that doubling the tamoxifen dose to 40mg/day, which may have normalized impaired metabolism, was clearly associated with an increased concentration of endoxifen in IMs [27,42,43,58,59]. Nevertheless, endoxifen plasma levels remained considerably low in PMs taking 40 mg/day [27] and several studies showed that CYP2D6 metabolizer status was not a reliable predictor of individualizing tamoxifen optimal dose [60]. Recently, it has been, however, found that adjusted doses of tamoxifen to 80 and 40 mg for poor and intermediate CYP2D6 metabolizers, respectively, had achieved therapeutic endoxifen levels in approximately 90% of patients. However, inter-patient variability remained [61-63]. Thus, it is still debatable whether patients benefit from the dose escalation of tamoxifen. Interestingly, recent data showed that the PM status did not affect the clinical outcomes in majority of the patients [64]. Furthermore, studies showed that CYP2D6 genotype alone can only partly explain the interindividual variability among patients treated with tamoxifen and that tamoxifen treatment cannot be individualized only by CYP2D6 genotype suggesting other genes involved in the metabolism of tamoxifen should be considered [30,65-69]. Therefore, a definitive exposureresponse effect remains debated and additional research is needed to determine whether CYP2D6 PMs phenotypes or even patients with low endoxifen plasma levels will achieve better clinical outcomes with increased dose of tamoxifen.
In spite of the fact that CYP2D6 genotyping test for tamoxifen treatment might be appropriate to optimize treatment outcome in specific patients [70], at present, it is still not generally recommended. The analysis of the existing literature and current evidence seem to be insufficient to justify the clinical utilization of CYP2D6 pharmacogenetic test to effectively guide dose and decision making in women with breast cancer for whom tamoxifen is being considered. Hence, it seems to be immensely challenging to reach consensus whether tamoxifen treatment should be based on the CYP2D6 testing and whether CYP2D6 genotyping approach is reliable and accurate in predicting treatment outcomes. For reliable implementation of CYP2D6 genotyping, large prospective studies of uniformly-treated women which establish evidence that the predictive capability of CYP2D6 testing exceeds the current approaches are required. Strategies such as therapeutic drug monitoring (TDM) and approaches based on phenotyping or direct measurements of endoxifen concentrations or even more recently administering chemical compounds that are unaffected by CYP2D6 metabolism such as Z-endoxifen have proved to be more useful and effective than CYP2D6 genotyping to individualize tamoxifen therapy. However, their value must be validated in prospective clinical studies as well [63,70-73].
Discussion
Recommendations and clinical guidelines
American Society of Clinical Oncology clinical practice (ASCO) guidelines and the National Comprehensive Cancer Network (NCCN) recommendations: Because of the controversial findings from the two above-mentioned big studies, BIG 1–98 and ATAC trial, in which no relationship was found between the CYP2D6 genotype and breast cancer recurrence, the committee of the American Society of Clinical Oncology clinical practice (ASCO) and the National Comprehensive Cancer Network (NCCN) do not recommend CYP2D6 genotype testing to personalize tamoxifen therapy and therefore CYP2D6 allelic status should not be used to guide adjuvant endocrine therapy. Although no definitive conclusion was made with regard to the interaction between CYP2D6 inhibitors and tamoxifen, NCCN and ASCO recommend that concomitant use of CYP2D6 inhibitors and tamoxifen should be avoided due to drug-drug interactions [74-76].
The FDA recommendations: Initial findings from retrospective studies prompted the FDA Advisory Committee in 2006 to recommend re-labelling tamoxifen to state that PMs patients have a high incidence of recurrence in comparison with other genotypes [34,77]. Since then, several studies have reported conflicting and highly discordant results [7,44,70,78]. At present, the FDA does not recommend the pharmacogenetic testing for CYP2D6 and current FDA label of tamoxifen does not refer to CYP2D6 genotyping [79,80].
PharmaGKB and the Dutch pharmacogenetics working group recommendations: For CYP2D6 poor (PMs) and intermediate (IMs) metabolizers, alternative drugs are recommended for treating postmenopausal patients with breast cancer and concurrent use of CYP2D6 inhibitors with tamoxifen to be avoided [81].
Conclusion
As genotyping costs drop and CYP2D6 genotyping has the potential to guide hormone therapy, one may inevitably anticipate a wide adoption of the test in clinical setting. However, the existing evidence does not support a statistically strong correlation between variant genotypes of CYP2D6 and breast cancer outcomes. The discrepant and conflicting findings regarding the influence of CYP2D6 genotype on clinical outcomes with tamoxifen have caused perplexity among regulatory bodies and health professionals.
Even if the evidence was consistent for a strong association between CYP2D6 genotype and treatment outcomes, at best, reimbursement decisions by governmental or insurance schemes are based partly on evidence of cost-effectiveness. Therefore, the major challenges that are hindering the adoption of CYP2D6 genotyping include the relatively small proportion of non-response to tamoxifen, cost implications of testing and availability of alternative drugs that may exceed the benefits achieved by tamoxifen therapy. One could argue that it is unethical not to offer CYP2D6 pharmacogenetic screening prior to initiation of tamoxifen to women who might take it for 5 or 10 years, since it is known that certain women indeed will not respond to a standard dose of it. However, to achieve convincing evidence of clinical validity, large scale prospective randomized trials are warranted to robustly and quantitatively demonstrate that knowledge of the patient’s CYP2D6 genotype can improve overall survival or reduce overall treatment cost in clinical practice. Further functional analysis is also required to elucidate the biological mechanisms underlying the metabolism and efficacy of tamoxifen and to further our understanding of the association between CYP2D6 phenotype and long-term clinical outcome.
References
- Howell A (2005) Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years' adjuvant treatment for breast cancer. Lancet 365: 60.
- Osborne CK (1998) Tamoxifen in the treatment of breast cancer. N Engl J Med 339: 1609-1618.
- Rolla R, Vidali M, Meola S, Pollarolo P, Fanello MR, et al. (2011) Side effects associated with ultrarapid cytochrome P450 2D6 genotype among women with early stage breast cancer treated with tamoxifen. Clinical Laboratory 58: 1211-1218.
- Veronesi U, Maisonneuve P, Rotmensz N, Costa A, Sacchini V, et al. (2003) Italian randomized trial among women with hysterectomy: tamoxifen and hormone-dependent breast cancer in high-risk women. J Natl Cancer Inst 95: 160-165.
- Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, et al. (1998) Tamoxifen for prevention of breast cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst 90: 1371-1388.
- Samer CF, Lorenzini KI, Rollason V, Daali Y, Desmeules JA (2013) Applications of CYP450 testing in the clinical setting. Mol diagn ther 17: 165-184.
- Regan MM, Leyland-Jones B, Bouzyk M, Pagani O, Tang W, et al. (2012) CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the breast international group 1-98 trial. J Natl Cancer Inst 104: 441-451.
- Moyer AM, Suman VJ, Weinshilboum RM, Avula R, Black JL, et al. (2011) SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics 12: 1535-1543.
- Kiyotani K, Mushiroda T, Tsunoda T, Morizono T, Hosono N, et al. (2012) A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Human Mol Genetics 21: 1665-1672.
- Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, et al. (2010) Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol 28: 1287-1293.
- Jordan VC, Collins MM, Rowsby L, Prestwich G (1977) A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol 75: 305-316.
- Crewe HK, Ellis SW, Lennard MS, Tucker GT (1997) Variable contribution of cytochromes P450 2D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem Pharmacol 53: 171-178.
- Desta Z, Ward BA, Soukhova NV, Flockhart DA (2004) Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: Prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther 310: 1062-1075.
- Jordan VC (1982) Metabolites of tamoxifen in animals and man: Identification, pharmacology, and significance. Breast Cancer Res Treat 2: 123-138.
- Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, et al. (2004) Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat 85: 151-159.
- Jin Y, Desta Z, Stearns V, Ward B, Ho H, et al. (2005) CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment J Natl Cancer Inst 97: 30-39.
- Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, et al. (2003) Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst 95: 1758-1764.
- Mürdter TE, Schroth W, Bacchusâ€ÂÂÂÂGerybadze L, Winter S, Heinkele G, et al. (2011) Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther 89: 708-717.
- Dehal SS, Kupfer D (1997) CYP2D6 catalyzes tamoxifen 4-hydroxylation in human liver. Cancer Res 57: 3402-3406.
- Lim HS, Lee JH, Lee SK, Lee SE, Jang IJ, et al. (2007) Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol 25: 3837-3845.
- https://www.cypalleles.ki.se/cyp2d6.htm.
- https://www.ncbi.nlm.nih.gov/books/NBK247013/.
- Sanderson S, Zimmern R, Kroese M, Higgins J, Patch C, et al. (2005) How can the evaluation of genetic tests be enhanced? Lessons learned from the ACCE framework and evaluating genetic tests in the United Kingdom. Genet Med 7: 495-500.
- Zanger UM, Raimundo S, Eichelbaum M (2004) Cytochrome P450 2D6: Overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol 369: 23-37.
- Bratherton DG, Brown CH, Buchanan R, Hall V, Pillers EK, et al. (1984) A comparison of two doses of tamoxifen (Nolvadex) in postmenopausal women with advanced breast cancer: 10 mg bd versus 20 mg bd. Br J Cancer 50: 199.
- Borges S, Desta Z, Li L, Skaar TC, Ward BA, et al. (2006) Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther 80: 61-74.
- Jr WJ, Walko CM, Weck KE, Ibrahim JG, Chiu WK, et al. (2011) Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: A multicenter study. J Clin Oncol 29: 3232-3239.
- Lash TL, Lien EA, Sørensen HT, Hamilton-Dutoit S (2009) Genotype-guided tamoxifen therapy: Time to pause for reflection?. Lancet Oncol 10: 825-833.
- Lyon E, Foster JG, Palomaki GE, Pratt VM, Reynolds K, et al. (2012) Laboratory testing of CYP2D6 alleles in relation to tamoxifen therapy. Genet Med 14: 990-1000.
- Charoenchokthavee W, Areepium N, Panomvana D, Sriuranpong V (2017) Effects of CYP2D6 and CYP3A5 polymorphisms on tamoxifen and its metabolites in Thai breast cancer patients. Breast Cancer: Targets and Therapy 9: 249.
- Hertz DL, Rae JM (2016) One step at a time: CYP2D6 guided tamoxifen treatment awaits convincing evidence of clinical validity. Pharmacogenomics 17: 823-826.
- Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, et al. (2005) Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 23: 9312-9318.
- Kiyotani K, Mushiroda T, Sasa M, Bando Y, Sumitomo I, et al. (2008) Impact of CYP2D6* 10 on recurrenceâ€ÂÂÂÂfree survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci 99: 995-999.
- Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, et al. (2007) Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol 25: 5187-5193.
- Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, et al. (2009) Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 302: 1429-1436.
- Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, et al. (2007) The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat 101: 113-121.
- Bonanni B, Macis D, Maisonneuve P, Johansson HA, Gucciardo G, et al. (2006) Polymorphism in the CYP2D6 tamoxifen-metabolizing gene influences clinical effect but not hot flashes: data from the Italian Tamoxifen Trial. J Clin Oncol 24: 3708-3709.
- Zembutsu H, Nakamura S, Akashi-Tanaka S, Kuwayama T, Watanabe C, et al. (2016) Association between CYP2D6 genotype and response to tamoxifen in a prospective multicenter study in Japan. Am Assoc Cancer Res 76: 2031-2031.
- Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, et al. (2005) Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat 91: 249-258.
- Gnant M, Mlineritsch B, Stoeger H, Luschin-Ebengreuth G, Poestlberger S, et al. (2010) Mature results from ABCSG-12: Adjuvant ovarian suppression combined with tamoxifen or anastrozole, alone or in combination with zoledronic acid, in premenopausal women with endocrine-responsive early breast cancer. J Clin Oncol 28: 533.
- Hertz DL, McLeod HL, Irvin WJ (2012) Tamoxifen and CYP2D6: A contradiction of data. The oncologist 17: 620-630.
- Wegman P, Vainikka L, Stål O, Nordenskjöld B, Skoog L, et al. (2005) Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res 7: R284.
- Wegman P, Elingarami S, Carstensen J, Stål O, Nordenskjöld B, et al. (2007) Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res 9: R7.
- Rae JM, Drury S, Hayes DF, Stearns V, Thibert JN, et al. (2012) CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst 104: 452-460.
- Zembutsu H, Nakamura S, Akashi-Tanaka S, Kuwayama T, Watanabe C, et al. (2016) Significant Effect of Polymorphisms in CYP2D6 on Response to Tamoxifen Therapy for Breast Cancer: A Prospective Multicenter Study. Clin Cancer Res 23: 2019-2026.
- Brauch H, Schroth W, Goetz MP, Mürdter TE, Winter S, et al. (2012) Tamoxifen use in postmenopausal breast cancer: CYP2D6 matters. J Clin Oncol 31: 176-180.
- Goetz MP, Ratain M, Ingle JN (2016) Providing balance in ASCO clinical practice guidelines: CYP2D6 genotyping and tamoxifen efficacy. J Clin Oncol 34: 3944-3945.
- Johnson JA, Hamadeh IS, Langaee TY (2015) Loss of heterozygosity at the CYP2D6 locus in breast cancer: implications for tamoxifen pharmacogenetic studies. J Natl Cancer Inst 107.
- Del Re M, Rofi E, Citi V, Fidilio L, Danesi R (2017) Should CYP2D6 be genotyped when treating with tamoxifen? Pharmacogenomics 18: 755-756.
- Ahern TP, Hertz DL, Damkier P, Ejlertsen B, Hamilton-Dutoit SJ, et al. (2016) Abstract P3-07-23: CYP2D6 genotype and breast cancer recurrence in tamoxifen treated patients: An evaluation of the importance of loss-of-heterozygosity. American Association for Cancer Research 76: 07-23.
- Markkula A, Hjertberg M, Rose C, Ingvar C, Jernström H (2014) No association found between CYP2D6 genotype and early breast cancer events in tamoxifen-treated patients. Acta Oncologica 53: 195-200.
- Park IH, Ro J, Park S, Lim HS, Lee KS, et al. (2012) Lack of any association between functionally significant CYP2D6 polymorphisms and clinical outcomes in early breast cancer patients receiving adjuvant tamoxifen treatment. Breast Cancer Res Treat 131: 455-461.
- Dezentje VO, Van Schaik RH, Vletter-Bogaartz JM, Van der Straaten T, Wessels JA, et al. (2013) CYP2D6 genotype in relation to tamoxifen efficacy in a Dutch cohort of the tamoxifen exemestane adjuvant multinational (TEAM) trial. Breast Cancer Res Treat 140: 363-373.
- Kidwell KM, Hertz DL, Leyland-Jones B, Regan MM, Dowsett M, et al. (2016) Abstract P6-09-02: Analysis of the International tamoxifen pharmacogenomics consortium (ITPC) dataset shows that genotyping DNA derived from tumor does not introduce CYP2D6 genotyping error or mask an association with tamoxifen efficacy. Cancer Res 76.
- Park GC, Jung JA, Bae KS, Lim HS (2017) A simulation study to compare the treatment effect of tamoxifen by CYP2D6 genotypes and thirdâ€ÂÂÂÂgeneration aromatase inhibitors. J Clin Pharmacol 57: 1088-1096.
- Yang Y, Botton MR, Scott ER, Scott SA (2017) Sequencing the CYP2D6 gene: From variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics 18: 673-685.
- Wickramage I, Tennekoon KH, Ariyaratne MA, Hewage AS, Sundralingam T (2017) CYP2D6 polymorphisms may predict occurrence of adverse effects to tamoxifen: a preliminary retrospective study. Breast Cancer Res Treat 9: 111.
- Kiyotani K, Mushiroda T, Imamura CK, Tanigawara Y, Hosono N, et al. (2012) Dose-adjustment study of tamoxifen based on CYP2D6 genotypes in Japanese breast cancer patients. Breast Cancer Res Treat 131: 137-145.
- Hertz DL, Deal A, Ibrahim JG, Walko CM, Weck KE, et al. (2016) Tamoxifen dose escalation in patients with diminished CYP2D6 activity normalizes endoxifen concentrations without increasing toxicity. Oncologist 21: 795-803.
- Fox P, Balleine RL, Lee C, Gao B, Balakrishnar B, et al. (2016) Dose escalation of tamoxifen in patients with low endoxifen level: evidence for therapeutic drug monitoring—the TADE Study. Clin Cancer Res 22: 3164-3171.
- Hertz DL, Rae JM (2016) Individualized tamoxifen dose escalation: Confirmation of feasibility, question of utility. Clin Cancer Res 22: 3121-3123.
- Klopp-Schulze L, Joerger M, Wicha SG, Ter Heine R, Csajka C, et al. (2017) Exploiting pharmacokinetic models of tamoxifen and endoxifen to identify factors causing subtherapeutic concentrations in breast cancer patients. Clin Pharmacokinetics 24: 1-4.
- Hennig EE, Piatkowska M, Karczmarski J, Goryca K, Brewczynska E, et al. (2015) Limited predictive value of achieving beneficial plasma (Z)-endoxifen threshold level by CYP2D6 genotyping in tamoxifen-treated Polish women with breast cancer. BMC Cancer 15: 570.
- Hwang GS, Bhat R, Crutchley RD, Trivedi MV (2017) Impact of CYP2D6 polymorphisms on endoxifen concentrations and breast cancer outcomes. The pharmacogenomics Journal.
- Spitman AS, Moes DJ, Gelderblom H, Dezentje VO, Swen JJ, et al. (2017) Effect of CYP3A4* 22, CYP3A5* 3, and CYP3A combined genotypes on tamoxifen metabolism. Eur J Clin Pharmacol.
- Sutiman N, Lim JS, Muerdter TE, Singh O, Cheung YB, et al. (2016) Pharmacogenetics of UGT1A4, UGT2B7 and UGT2B15 and their influence on tamoxifen disposition in Asian breast cancer patients. Clinical Pharmacokinetics 55: 1239-1250.
- Schroth W, Winter S, Mürdter T, Schaeffeler E, Eccles D, et al. (2017) Improved prediction of endoxifen metabolism by CYP2D6 genotype in breast cancer patients treated with tamoxifen. Front Pharmacol 8: 582.
- Bertholee D, Maring JG, Van Kuilenburg AB (2017) Genotypes affecting the pharmacokinetics of anticancer drugs. Clinical Pharmacokinetics 56: 317-337.
- Novillo A, Romero-Lorca A, Gaibar M, Rubio M, Fernández-Santander A (2017) Tamoxifen metabolism in breast cancer treatment: Taking the focus off the CYP2D6 gene. Pharmacogenomics Journal 17: 109-111.
- Del Re M, Citi V, Crucitta S, Rofi E, Belcari F, et al. (2016) Pharmacogenetics of CYP2D6 and tamoxifen therapy: Light at the end of the tunnel?. Pharmacological Res 107:398-406.
- Binkhorst L, Mathijssen RH, Jager A, Van Gelder T (2015) Individualization of tamoxifen therapy: much more than just CYP2D6 genotyping. Cancer Treat Rev 41: 289-299.
- Goetz MP, Suman VJ, Reid JM, Northfelt DW, Mahr MA, et al. (2017) First-in-human phase I study of the tamoxifen metabolite Z-endoxifen in women with endocrine-refractory metastatic breast cancer. J Clin Oncol 30: JCO2017733246.
- Koolen SL, Bins S, Mathijssen RH (2016) Individualized tamoxifen dose escalation. Clin Cancer Res 22: 6300-.
- Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, et al. (2016) Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol 34: 1134-1150.
- Gradishar WJ, Anderson BO, Blair SL, Burstein HJ, Cyr A, et al. (2014) Breast cancer version 3.2014J Natl Compr Canc Netw 12: 542-590.
- https://www.jnccn.org/content/15/9/1140.full.
- https://www.fda.gov/ohrms/dockets/ac/06/briefing/2006-4248B1-01-FDA-Tamoxifen%20Background%20Summary%20Final.pdf.
- Damkier P, Kjærsgaard A, Barker KA, Cronin-Fenton D, Crawford A, et al. (2017) CYP2C19* 2 and CYP2C19* 17 variants and effect of tamoxifen on breast cancer recurrence: Analysis of the International Tamoxifen Pharmacogenomics Consortium dataset. Scientific Reports 7: 7727.
- https://www.accessdata.fda.gov/drugsatfda_docs/label/2005/17970s053lbl.pdf.
- https://www.fda.gov/Drugs/ScienceResearch/ucm572698.htm.
- https://www.pharmgkb.org/gene/PA128/guideline.


