Keywords
Acquired immunodeficiency syndrome; Protease inhibitor; Genetics
Introduction
Since 1995, Anti-Retroviral Therapy (ART) has been widely and extensively used to treat HIV infection and slow progression toward Acquired Immunodeficiency Syndrome (AIDS). ART comprises a combination of drugs that target different steps of Human Immunodeficiency Virus (HIV) replication to reduce viral load, minimize opportunistic infections, allow immune reconstitution, and ultimately lower the mortality rate of AIDS and associated diseases [1,2]. However, HIV-1 evolves rapidly and ART can lead to acquired mutations that confer drug resistance. Importantly, transmission of resistant HIV-1 strains may cause severe clinical and epidemiological problems, while therapeutic failure resulting from drug resistant HIV-1 strains has become a major factor that limits successful antiviral treatments [3,4]. As such, surveillance of HIV-1 mutations that do not respond to ART may provide additional information that could guide novel drug development and treatment strategies. The present study investigated ART-virological failure prevalence, mutation variations, and genetic trends exhibited by HIV-1 strains isolated from HIV-seropositive patients in Hebei province, China that underwent antiviral treatment between 2008 and 2013.
Materials and Method
Patient cohort
A total of 569 HIV-seropositive and AIDS patients (551 adults and 11 children) that received antiviral treatment between 2008 and 2013 at a disease prevention and control center in Hebei, China were included in this study. No patients were included for 2009 because of a lack of participation.
An EDTA anti-coagulated whole blood sample (10 ml) was collected from each patient. Fifty microliters of each sample were analyzed for CD4 cell counts (FACSCount, Becton Dickinson, USA). Plasma was separated from the remaining blood sample and analyzed for HIV-1 viral loads (VL; COBAS AMPLlCOR, Roche, USA). A VL ≥ 1000 copies/ml was the criterion used for treatment failure.
HIV-1 RNA isolation and genotyping
HIV-1 RNA was isolated from each plasma sample with a QIAamp ®Viral RNA Mini isolation kit (QIAGEN, Hilden, Germany). The pol gene sequence was amplified twice by Polymerase Chain Reaction (PCR) with primers [5]. PCR products were analyzed by electrophoresis on a 1% (w/v) agarose gel. PCR products were sequenced by Sangon Biotech Co. Ltd. (Shanghai, China). Sequence fragments were assembled with Contig Express (https://www.lifetechnologies.com/ca/en/home/technicalresources/ order-support.html), compared with the sequencing chromatogram, and analyzed for drug resistant mutations using the HIV Drug Resistance Database at Stanford University, CA, USA (hivdb.stanford.edu) [6].
Statistical analysis
Data were analyzed with the χ2-test, the χ2-test for trends, and the Fisher’s exact probability test using SPSS 17.0 software (SPSS, Chicago, USA). A p value<0.05 was considered statistically significant.
Ethics statement
All investigations and HIV-1 tests were informed and voluntary, and participants agreed that their samples may be used for the purpose of controlling and preventing HIV. Written informed consent was obtained from all adult patients and parents/ guardians of HIV-1 positive children in this study. Our study was approved by the local Ethics Committee at Hebei Province Centers for Disease Control and Prevention (IRB-2012004).
Results
Prevalence of HIV-1 drug resistance
Between 2008 and 2013, the percentage of patients with drug resistance dropped from 60.9% to 35.0% among ART-failure patients, and this decreasing trend was statistically significant (p<0.05). However, when all HIV-positive patients undergoing antiviral treatment (including ART success patients) were included in our analysis, differences in prevalence and trends were not statistically significant (Table 1, p>0.05).

Table 1: HIV-1 drug resistance among ART-failure patients between 2008 and 2013.
HIV-1 drug resistance among different antiretroviral regimens
As shown Table 2, the first line antiretroviral drugs were included in regimen 1, regimen 2 and other regimens. Regimen 1 and regimen 2 belonged to the first line therapeutic regimens, and other regimens contained LPV/r. The distribution of HIV-1 DR in these three regimens between 2008 and 2013 showed no significant difference (P>0.05) and no significant trend (χ2=0.088, P=0.766). However, in 2011, the distribution of HIV-1 DR in these three regimens showed a significant difference (P<0.05), and the proportion of HIV-1 DR in regimen 1(86.1%, 31/36) was obviously higher than regimen 2(4.3%, 3/7) and other regimens (4.0%, 2/5). There were no significant different distribution of DR in 2008(P>0.05), 2010(P>0.05), 2012(P>0.05) and 2013(χ2=2.232, P=0.328), respectively.
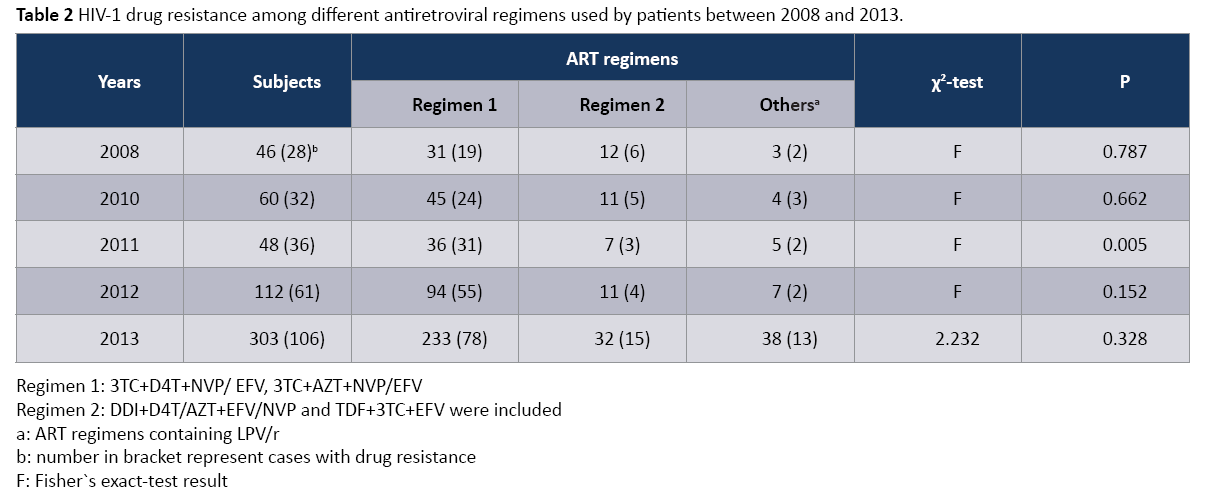
Table 2: HIV-1 drug resistance among different antiretroviral regimens used by patients between 2008 and 2013.
HIV-1 drug resistance among different HIV-1 genotypes
As shown in Table 3, HIV-1 genetic diversity was identified in this study, from subtype B and CRF01_AE in 2008 to subtype B, subtype C, CRF01_AE, CRF07_BC, CRF08_BC, CRF02_AG and URF. The distribution of HIV-1 DR in three main subtypes (subtype B, subtype C and CRF01_AE) showed the significant difference (P<0.001). Particularly, the proportion of HIV-1 DR in subtype B indicated the significant decreasing trend (χ2=98.571, P<0.001), from 2011 to 2013. The above phenomenon suggested that HIV- 1 genetic diversity and DR difference has become the barrier of HIV prevention.
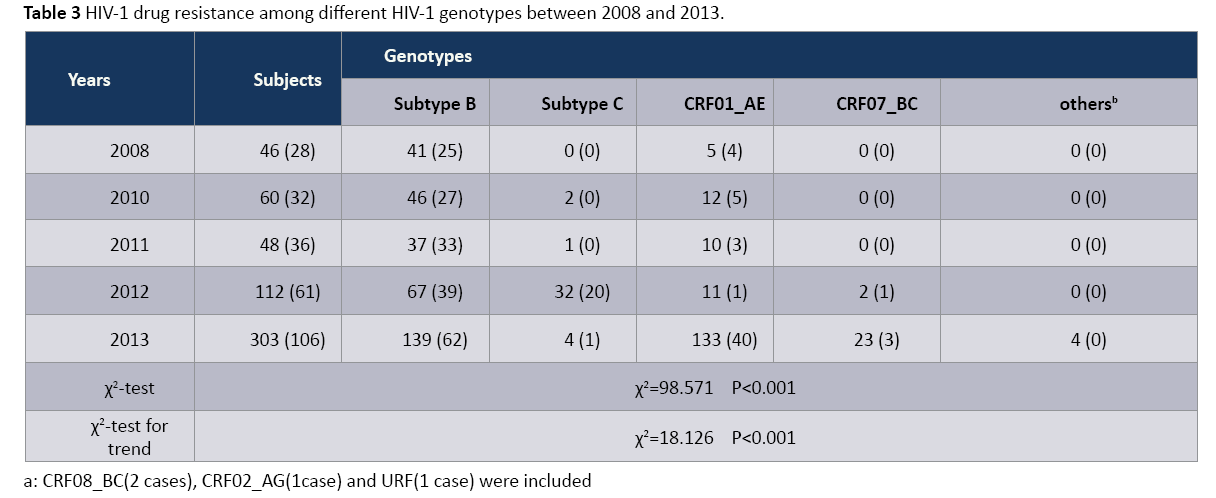
Table 3: HIV-1 drug resistance among different HIV-1 genotypes between 2008 and 2013.
HIV-1 RNA mutations: HIV-1 protease gene mutations
Mutations detected in the HIV-1 protease coding region were analyzed (Table 4). Six different mutations were detected in HIV- 1strains isolated from patients in 2010 (V32AV, Q58E, M46IM, M46L, L10I, A71V/T), three mutations were detected in HIV- 1 strains isolated from patients in both 2012 and 2013 (M46L, L10I, A71V/T), and two mutations were detected in HIV-1 strains isolated from patients in 2011 (L10I, A71V/T). Of these, only three mutations (M46IM, M46L, and Q58E) were related to Protease Inhibitor (PI) treatment failure. Conversely, the most frequently observed mutations in this patient cohort (A71V/T, L10I, 32AV) were not related to PI treatment failure. Only one mutation (A71V/T) rate displayed a significant difference among ARTfailure patients between 2010 and 2013 (p<0.05). When tested for trends, however, we did not detect any statistically significant trend for any of the six mutations that were identified (p>0.05).
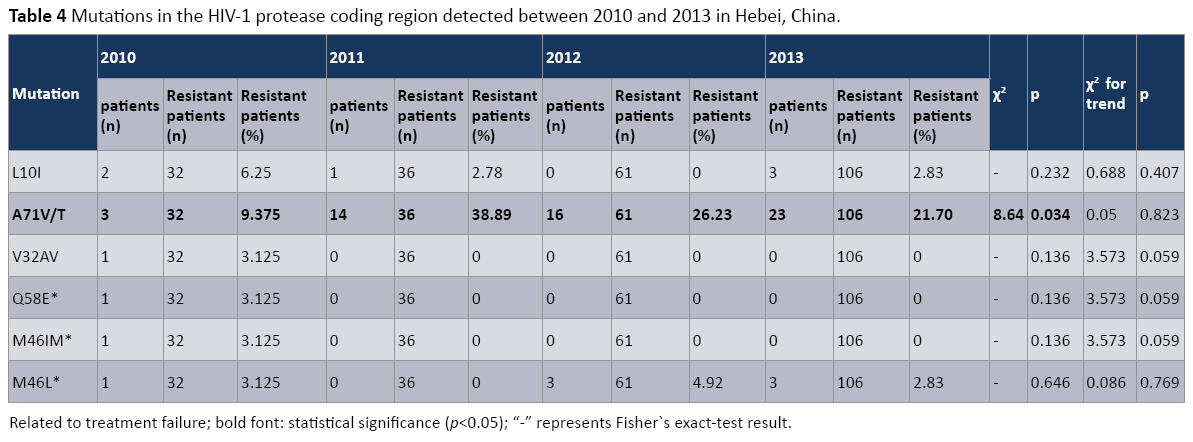
Table 4: Mutations in the HIV-1 protease coding region detected between 2010 and 2013 in Hebei, China.
HIV-1 reverse transcriptase gene mutations related to Nucleoside Reverse Transcriptase Inhibitors (NRTIs)
Tables 3 and 4 list detected mutations related to Reverse Transcriptase Inhibitor (RTI) treatment failure in the patient cohort. Between 2010 and 2013, 12 mutations related to Nucleoside Reverse Transcriptase Inhibitor (NRTI) treatment failure were found in the HIV-1 reverse transcriptase coding region: A62V, M184V, T215Y, M41L, L210W, D67N, K70R, T215F, K219E, K65R, Q151M, and L74V/L (Table 5). Of these, M184V occurred most frequently (68.75% in 2010 and 57.55% in 2013). Nine mutations (V75I, T215Y, M41L, L210W, T69D, D67DG, V118I, V75I/T, and F77L) exhibited statistically significant differences in prevalence during this period (p<0.05). The observed decreasing trend for all nine mutations was statistically significant (p<0.05). Additionally, two mutations (T215F and Q151M) exhibited no statistically significant changes during this time period; however, they both demonstrated decreasing trends that were significant (both p<0.05).
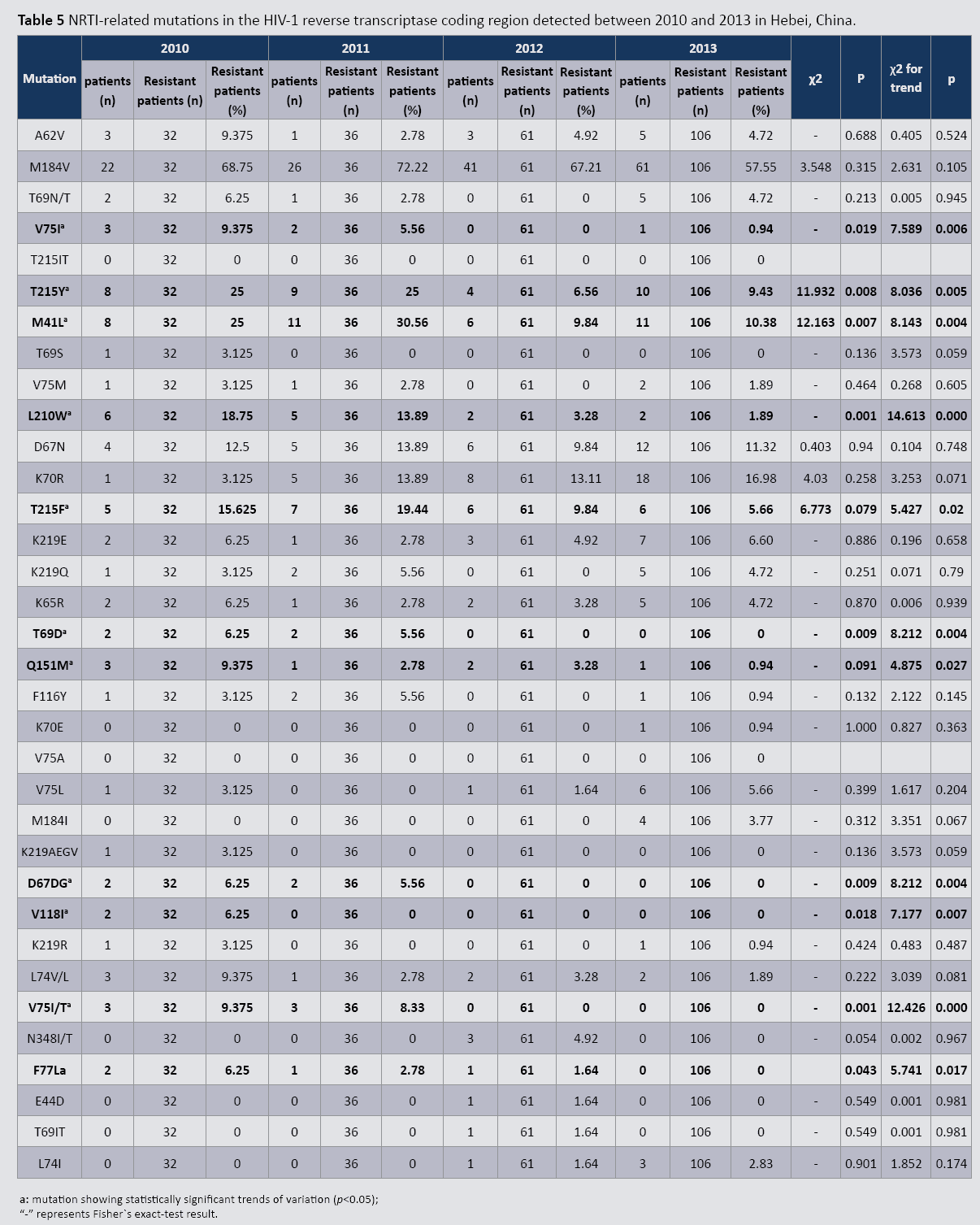
Table 5: NRTI-related mutations in the HIV-1 reverse transcriptase coding region detected between 2010 and 2013 in Hebei, China.
HIV-1 reverse transcriptase gene mutations related to non-nucleoside reverse transcriptase inhibitors (NNRTIs)
Of all NNRTI-related mutations (Table 6), K103N occurred most frequently each year between 2010 and 2013 (40.62%–25.47%). This was followed by Y181C (40.625–32.08%), and V108I (21.87%–7.55%). During this period, V108I and M230L displayed statistically significant decreasing trends, whereas K238T and V90I exhibited significant increasing trends (p<0.05). Furthermore, only V106I, M230L, and V179D exhibited statistically significant differences (p<0.04) in prevalence during this period.
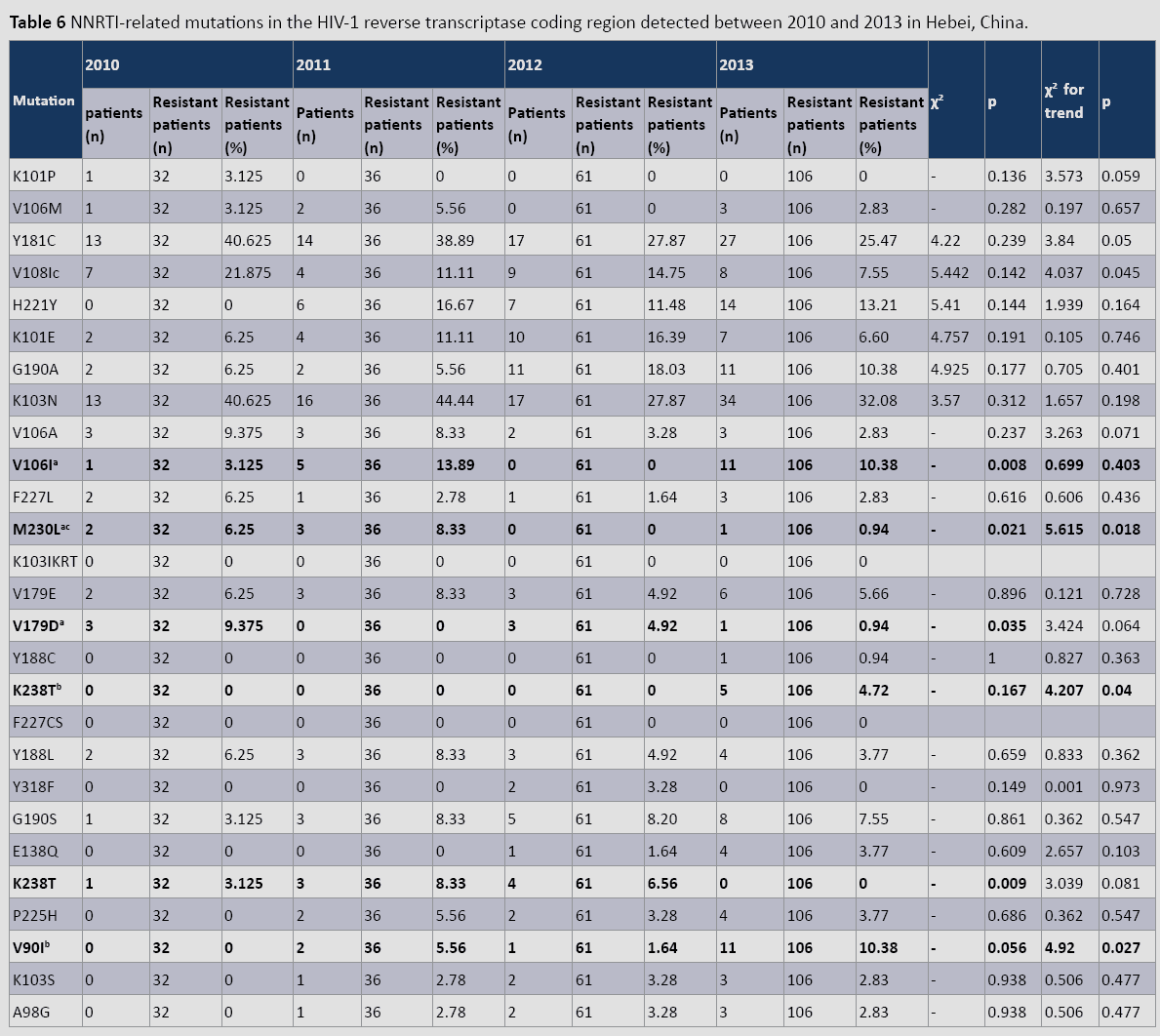
Table 6: NNRTI-related mutations in the HIV-1 reverse transcriptase coding region detected between 2010 and 2013 in Hebei, China.
Discussion
Since the appearance of the first anti-HIV agent, zidovudine, extensive and widespread use of antiviral therapies has induced adaptive gene mutations in HIV RNA. Li et al. reported that drugresistant HIV strains account for 43.14% of all investigated strains [6], while Boden et al. found that 5–16% of newly HIV-infected individuals in Europe and the USA are infected with HIV-1 drugresistant strains [7]. Epidemiological data have clearly linked drug resistance resulting from HIV-1 mutations to failed antiviral treatment and accelerated patient death [8,9].
Since the first HIV infection reported in Hebei Province in 1989, the cumulative 4148 HIV-1-infected individuals and 1860 AIDS patients have been reported in the province by the end of 2013,. The recorded mortality rate for these AIDS patients to date is approximately 31%. In 2003, the Chinese government issued a set of “free care and financial aid” policies regarding treatment of HIV-infected individuals and AIDS patients. These policies have substantially reduced morbidity and mortality in these groups, as well as improved quality of life.
It has been reported that during antiretroviral treatment, approximately 50% of HIV-infected patients develop at least one major drug-resistant viral mutation that greatly affects treatment outcome [10-15]. In 2007, Chen et al. studied drug resistance in AIDS patients undergoing antiviral treatment in Hebei, China [16]. They found that only 2.6% of AIDS patients in this Chinese province developed drug resistance, and this was significantly lower than comparable data recorded in other regions of the country [6,11,17,18]. In the present study, we report that 60.9% of patients with undergoing antiviral treatment failed to respond to treatment in 2008. We analyzed the change in drug-resistant mutations between 2008 and 2013, and found that the proportion of patients with treatment failure decreased significantly during this period. However, when all HIV-infected individuals were included in our analysis, no statistically significant trend was observed. Furthermore, the distribution of HIV-1 DR in these three regimens between 2008 and 2013 showed no significant difference and no significant trend. This study result suggested that drugs used for antiretroviral therapy with a long time were the main cause of HIV-1 DR, including poor drugadherence and no change of regimens. Our study also revealed HIV- 1 genetic diversity and DR difference among these subtypes, which has become the barrier of HIV prevention.
Taken together, these findings support a growing body of evidence demonstrating that drug resistance should be monitored frequently and treatment regimens be updated appropriately [19]. Both can potentially improve the therapeutic effects of antiviral treatments for people living with HIV and AIDS.
Author’s Contribution
HZ and XL implemented the study, analyzed results and drafted the manuscript. XL contributed to the acquisition of the data and revising it critically. YZ and GB carried out the epidemiological investigation. WW performed the statistical analysis. CZ, YL and YW participated in the experiment of HIV-1 molecular biology.
Acknowledgement
We thank Academy of Military Medical Sciences for technical assistance in this study. This work was supported by Research Fund Project of Hebei Province for Health (ZL20140045) and Scientific and Technological Research Plan of Hebei Province (13277716D).
References
- Looney D, Ma A, Johns S (2015) HIV Therapy-The State of ART. Curr Top Microbiol Immunol 389:1-29.
- Mukherjee A, Shah N, Singh R, Vajpayee M, Kabra SK, Lodha R (2014) Outcome of highly active antiretroviral therapy in HIV- infected Indian children. BMC Infect Dis 14:701.
- Zhang F, Dou Z, Ma Y, Zhao Y, Liu Z, et al. (2009) Five-year outcomes of the China National Free Antiretroviral Treatment Program. Ann Intern Med 151:241–251.
- Yang WL, Kouyos RD, Böni J, Yerly S, Klimkait T,et al. (2015) Persistence of Transmitted HIV-1 Drug Resistance Mutations Associated with Fitness Costs and Viral Genetic Backgrounds. PLoS Pathog 11:e1004722.
- Li H, Zhong M, Guo W, Zhuang D, Li L, Liu Y, et al.(2013)Prevalence and mutation patterns of HIV drug resistance from 2010 to 2011 among ART-failure individuals in the Yunnan Province, China. PLoS One 8:e72630.
- Li HP, Li H, Yang K, Wang Z, Bao ZY, et al. (2005) Research of clinically efficacy in patients treated with antivirus drug and the untreated controls at partial region, Henan province and drug resistance. Chin J Microbio Immunol 25:194-198.
- Boden D, Hurley A, Zhang L, Cao Y, Guo Y, et al. (1999) HIV-1 drug resistance in newly infected individuals. JAMA 282:1177-1179.
- Jay A Levy (2000)HIV and pathogenesis of AIDS. ASM Science.
- Saenz RA, Bonhoeffer S (2013) Nested model reveals potential amplification of an HIV epidemic due to drug resistance. Epidemics 5:34-43.
- Susman E (2002) Many HIV patients carry mutated drug-resisitant strains. Lancet359:49-50.
- Yao X, Peng GP, Tang H, Jiang HL (2008) Drug resistance to antiretroviral therapy for HIV/AIDS patients. Chin J Lepr Skin Dis 24: 430-432.
- Li JY, Li H, Li HP (2005) Prevalence and evolutionary characteristics of drug-resistant HIV strain in Henan province, China.Bull Acad Military Med Sci 29:8-12.
- Lu XL, Zhao HR, Zhang YQ,Zhao CY, Li HP, et al. (2013) Drug resistance mutation of HIV-1 in HIV/AIDS patients infected by blood transfusion. Med J Chin PLA 38: 230-234.
- Zhao HR, Lu XL, Li Y, Bai GY, Wang YY, et al. (2013) Drug resistance variation in HIV-1 after the failure of highly active antiretroviral therapy. Chin J Infect Control 12:247-250.
- Zhao HR, Lu XL, Zhao CY, Li BJ, Chen ZQ, et al. (2012) Analysis on drug-resistance in 36 patients treated with antivirus-therapy in Hebei Province. Chin J Dis Control Prev16: 412-416.
- Chen ZQ, Liang L, Li BJ, Wang CY, Zhang CX, et al. (2009) HIV-1 Drug resistance among HIV/AIDS patients in Hebei Province in 2007. Hebei Med J31:1129-1130.
- Li HP, Liu W, Liu HX, Liang SJ, Bao ZY, et al. (2007) Study on the antiviral therapy program among people with human immunodeficiency virus in Guangxi Zhuang authority. Chin J Epidemiol 28: 338-343.
- Wu JJ, Xin H, Shen YL, Liao LJ, Su B, et al. (2011) Study on transmission of drug resistant human in immunodeficiency virus-1 naive infection in Anhui Province in 2008. Chin J Dis Control Prev 15: 33-35.
- Little SJ, Holte S, Routy JP, Eric S, Marty Markowitz , et al. (2002) Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med 347:385-394.