Research Article - (2023) Volume 9, Issue 4
Evaluation of the Antimicrobial Producing Actinomycetes from Regions in Baghdad City
Balqees Yahya Najm*,
Sarab Hussein Khallel and
Hala Mahmmed Majeed
Department of Basic Science, Ibn Sina University of Medical and Pharmaceutical, Khartoum, Sudan
*Correspondence:
Balqees Yahya Najm, Department of Basic Science, Ibn Sina University of Medical and Pharmaceutical,
Khartoum,
Sudan,
Email:
Received: 21-Oct-2022, Manuscript No. IPBMBJ-22-14570;
Editor assigned: 24-Oct-2022, Pre QC No. IPBMBJ-22-14570 (PQ);
Reviewed: 07-Nov-2022, QC No. IPBMBJ-22-14570;
Revised: 22-Aug-2023, Manuscript No. IPBMBJ-22-14570 (R);
Published:
29-Aug-2023, DOI: 10.36648/2471-8084.9.4.32
Abstract
The aim of the present study includes the isolation of 25 isolates belonging to the actinomycetes from soil samples collected from different agricultural regions in Baghdad city. The isolates were identified tentatively on the basis of chalky, leathery. Mucoidy, waxy, appearance of colonies. The identification was confirmed by using slide culture technique and observing substrate and aerial mycelium, and arrangement of spore chains. 10 isolates showed capability to produce antimicrobial activity. To more confirm that these isolates were belonged to actinomycetes, they were identified by molecular method using 16rDNA they were obtained a specific band of about 1500 bp the selected isolates were studied for their antimicrobial activity against gram positive and gram negative bacteria. Also study conducted studying the influence of factors such carbohydrate (glucose, maltose, sucrose, lactose and starch) and different concentration of (NH4)2SO4 as nitrogen source on antimicrobial production.
Keywords
Actinomycetes; Biosynthetic; Cluster gene; Actinomycetin; Proactinomycin; Streptomycin
Introduction
Microbial natural products are on the cutting edge of the search for bioactive molecules with medicinal potential. Actinomycetes are responsible for more than 80% of all naturally occurring antibiotics on the market. Actinomycetes are gram positive, filamentous, spore forming, aerobic bacteria that are found throughout the natural world. Their DNA has a high GC content (>55 mol%), and their spore producing ability and mycelia structure are unusual. Aerial hyphae produced by actinomycetes develop into spore chains. Many microorganisms, especially actinomycetes, rely on soil to produce a variety of bioactive natural resources, including clinically relevant medicines. Actinomycetes have more genes that codes for enzymes that help them produce bioactive secondary metabolites. According to the literature, strain growth circumstances influence secondary metabolite production, which is why researchers make nutritional and physiochemical changes throughout the fermentation process and use genome sequencing to identify genes involved in secondary metabolite production. The majority of bioactive compounds are obtained by metabolic pathways, which are encoded by genes on neighboring chromosomes (biosynthetic gene cluster). Multidrug resistance is currently a big problem, which is worsening due to a decrease in the supply of new antibiotics. Finding new bacteria and fungi isolation procedure as a result, research is now focused on finding next generation medicinal compounds that can be exploited in previously undiscovered actinomycetes environments. Actinomycetin, mycetin, and micromonosporin lysozyme, as well as actinomycin, streptothricin, proactinomycin, and streptomycin, are all significant antibiotics generated by actinomycetes. The structural, antibacterial, and cytotoxic properties of these antibiotics vary substantially. As well as searching for new places or environments for microbe selection, could lead to new antibiotics. However, in the last two decades, the number of new antibiotic compounds discovered has decreased. However, in the last two decades, the number of new antibiotic compounds discovered has decreased. As a result, research is currently focused on finding the next generation pharmaceutical compounds with never before seen homes actinomyces. Some of the most important antibiotic made by Actinomycetin, mycetin and micromonosporin are all actinomycetes actinomycin, streptoth ricin, proactinomycin, and lysozyme. The structure of these antibiotics varies widely characteristics antibacterial and toxicity. Streptomyces spp., including Streptomyces kanamyceticus, Streptomyces fradiae, Streptomyces griseus, Streptomyces antibioticus, Streptomyces venezuelae, Streptomyces lincolnensis, Streptomyces roseosporus, and Actinoplanin teichomyceticus, are the largest genus most commonly found in terrestrial habitat Streptomyces is known for producing a wide range of extracellular enzymes and bioactive secondary metabolites with a wide range of structural and functional diversity that are used as antibacterial, antiprotozoal, antifungal, antiviral, anthelminthic, anticholesterol, anticancer, immunosuppressant, pesticides, and herbicides. Bioactive secondary metabolites produced by Streptomyces have attracted a lot of attention because of their helpful biological activities, especially in terms of human consequences health a constant search for potential bacterial taxa. The creation of secondary metabolites is necessary for the development of new drugs. The goal of the present study was to evaluate actinomyces for their potential of secondary metabolites, and detection of antibacterial of local isolates against pathogenic bacteria [1-5].
Materials and Methods
Sample Collection
200 soil samples were collected from different regions of agricultural regions in Baghdad city starting from 3/3/2022 for one months, these samples are collected from landscape (6 cm-12 cm ) dept., these sample were dried and put in sterilized polyethylene zipper bags and they were stored at 5°C in refrigerated until used.
Isolation of Actinomycetes
Dilutions of soil samples were made by dissolving them in distilled water. For mycelia disruption, 1 percent sodium dodecyl sulfate SDS was added. Enrichment is necessary for the growth of actinomycetes. As antifungal agents, nistatin was added to the media L50 of each dilution ere speared on enrichment media plates and incubated at 31°C for 7 to 13 days.
Morphological Identification of Actinomyces
Actinomycetes strains were identified morphologically using traditional microscopy procedures and a variety of technique Magnification lenses ranging from 10 to 100 times magnification the morphology of these actinomyces was then compared to that of other actinomyces.
Molecular Characterization of Actionomyces
Actinomycetes were identified molecularly using Polymerase Chain Reaction (PCR) and 16S rRNA gene amplification techniques. The methodology for genomic Deoxyribo Nucleic Acid (DNA) isolation was followed. To amplify the fragment of 16S rDNA gene, a set of primers (i.e., forward (St-F): 5-AAGCCCTGGAAACGGGGT-3 and reverse (St-R): 5-CGTGTGCAGCCCAAGACA-3) was used. PCR amplification was performed using master mix (operon technologies), 0.4 μM primer, 40 ng chromosomal DNA, and the final volume was reached to 25 μl. The PCR amplification was performed using the thermal cycler program as follows: 94°C for 5 min as a primary denaturation step, 35 cycles of 94°C for 1 min, 57°C for 1 min, 72°C for 105 sec and final extension was 72°C for 10 min. The PCR products were visualized using gel electrophoresis on 1% agarose and compared with 10 kb DNA ladder.
Antibiotic Production in Liquid Cultures
Antibiotic production medium were prepared by solving 0.8 g of Nacl, 1 NH4Cl, 0.1 K2HPO4, 0.2 MgSO4.7H2O, 0.04 CaCl2, 10 glucose and 3 g of yeast extract were solved in 1 L of distle water, Each media was made in a 500 ml flask, kept at a pH of 7.2, and autoclaved for 20 minutes at 121°C and 15 pressure. Actinomycete spores were inoculated under sterile circumstances. The cultures were grown in a shaking incubator at 120 rpm, 30°C for 8 days in order to produce secondary metabolites. These tests were all conducted in triplicate [6-10].
Extraction of Antimicrobial and Study their Activity
Secondary metabolites were extracted by method described by Ilic, et al.
Tests for Antimicrobials
After the fermentation period, 1.5 ml of supernatant from the fermented flask was transferred into Eppendorf tubes for the antimicrobial assay. Nutrient agar media containing colonies of Escherichia coli, Staphylococcus aureus, Bacillus subtilis, and Pseudomonas aeruginosa were used to evaluate the antimicrobial activity of actinomycetes.
Antibiotic Bioassay
Mueller-Hinton agar was used as the test medium, and the paper disc diffusion technique was used to carry it out. Mueller Hinton agar (45°C) was added and allowed to harden in sterile petri plates (9 cm in diameter). Agar surface was infected with 0.1 ml of the test organism's bacterial solution (3 x 106 cells). On the dry surface of the medium, sterile paper discs (6.0 mm in diameter, Whatman antibiotic test discs) were positioned. The culture filtrate was divided into 20 l portions for each disc. For two hours, Petri plates were placed in the refrigerator to allow the antibiotic to diffuse. The diameter of the inhibitory zone was measured in mm.
Study of Influence of Factors on Antimicrobial Production
This study also investigated study some factors and optimal conditions for antimicrobial production for one selected isolates. The carbon sources (glucose, malto In the basal medium, which contains K2HPO4, 1.0 g, MgSO4.7H2O, 0.5 g, CaCl2. 2H2O, 0.04 g, FeSO4.7H2O, 0.005 g, ZnSO4.7H2O, 0.0005 g, and water, 1,000 ml, carbon and nitrogen sources were examined. A pH adjustment of 7.5 was made sucrose, lactose and starch) at 1% concentration, and (NH4)2SO4 at concentration (0.05, 0.10, 0.15, and 0.20) were sterilized separately and added just prior to inoculation. These were added to the basic medium, which had its pH set at 7.2 and been sterilized 250 ml of Erlenmeyer flask are inoculated with 10 ml of Streptomyces inoculum. For each test, triplicate flasks were typically utilized. A rotary shaker was used to maintain the flasks (220 rpm) and seven days of incubation. Following that, a disc diffusion technique was developed to assess the antibacterial activity [11-15].
Results and Discussion
This study includes isolation of 25 (12%) isolates belonged to the genus Streptomyces spp. from 200 different rizosphere soil agricultural region in Baghdad city, these isolates were identified on the basis of chalky appearance of colonies and production of moist earthy odor then confirmed by using slide culture technique. Spore color, aerial and substrate mycelium formation, and production of diffusible pigment. Morphological examination of these isolates indicates that they belong to the genus Streptomyces (Table 1).
| Isolate no. |
Medium |
Growth |
Areal mycelium |
Substrate mycelium |
Diffusible pigment |
| Strp 1 |
GAA/SMSA/PGYEA/TA |
Good/moderate/good/good |
Gray/white/gray/brown |
White/brown/red/brown |
None/gray/red/white |
| Strp 2 |
GAA/SMSA/PGYEA/TA |
Good/good/good/moderate |
White/brown/none/gray |
Green/red/red/gray |
Brown/gray/white/red |
| Strp 3 |
GAA/SMSA/PGYEA/TA |
Moderate/good/good/moderate |
Red/white/white/red |
None/gray/none/red |
Gray/red/red/white |
| Strp 4 |
GAA/SMSA/PGYEA/TA |
Good/good/moderate/moderate |
Brown/gray/white/none |
Gray/white/white/white |
Brown/white/white/none |
| Strp 5 |
GAA/SMSA/PGYEA/TA |
Moderate/good/moderate/good |
Brown/gray/white/red |
Brown/gray/white/red |
Brown/gray/white/red |
| Strp 6 |
GAA/SMSA/PGYEA/TA |
Good/moderate/good/good |
Gray/red/red/white |
None/gray/none/red |
Gray/red/red/white |
| Strp 7 |
GAA/SMSA/PGYEA/TA |
Moderate/good/good/good |
Gray/gray/white/white |
None/gray/none/red |
None/brown/red/brown |
| Strp 8 |
GAA/SMSA/PGYEA/TA |
Moderate/good/good/moderate |
Gray/none/none/red |
White/brown/none/white |
Gray/red/red/white |
| Strp 9 |
GAA/SMSA/PGYEA/TA |
Moderate/good/moderate/good |
None/gray/none/red |
Gray/red/red/white |
Brown/brown/none/brown |
| Strp 10 |
GAA/SMSA/PGYEA/TA |
Good/good/moderate/good |
None/gray/none/red |
Gray/red/red/white |
Brown/gray/white/red |
| Strp 11 |
GAA/SMSA/PGYEA/TA |
Moderate/good/good/moderate |
White/brown/none/white |
Brown/brown/none/none |
Gray/red/red/white |
| Strp 12 |
GAA/SMSA/PGYEA/TA |
Good/good/moderate/moderate |
None/gray/none/red |
Gray/white/white/white |
Gray/red/red/white |
| Strp 13 |
GAA/SMSA/PGYEA/TA |
Good/moderate/good/good |
Gray/white/white/white |
None/gray/none/red |
Brown/gray/white/red |
| Strp 14 |
GAA/SMSA/PGYEA/TA |
Good/good/moderate/good |
Brown/gray/white/red |
Brown/brown/none/none |
Brown/brown/none/none |
| Strp15 |
GAA/SMSA/PGYEA/TA |
Good/moderate/good/good |
Gray/white/white/white |
None/gray/none/red |
Brown/gray/white/red |
| Strp 16 |
GAA/SMSA/PGYEA/TA |
Moderate/good/good/moderate |
Brown/brown/none/none |
None/brown/none/brown |
White/brown/none/white |
| Strp 17 |
GAA/SMSA/PGYEA/TA |
Good/moderate/good/good |
Gray/white/white/white |
Brown/white/white/none |
Brown/white/white/none |
| Strp 18 |
GAA/SMSA/PGYEA/TA |
Good/moderate/good/good |
White/brown/none/white |
Brown/brown/none/none |
Brown/gray/white/red |
| Strp 19 |
GAA/SMSA/PGYEA/TA |
Moderate/good/good/good |
Gray/white/white/white |
Brown/gray/white/red |
White/brown/none/white |
| Strp 20 |
GAA/SMSA/PGYEA/TA |
Good/moderate/ good/good |
Gray/red/red/white |
Brown/brown/none/none |
Gray/red/red/white |
| Strp 21 |
GAA/SMSA/PGYEA/TA |
Good/good/moderate/good |
Brown/brown/none/none |
Gray/red/red/white |
Brown/gray/white/red |
| Strp 22 |
GAA/SMSA/PGYEA/TA |
Good/moderate/good/good |
Gray/white/white/white |
White/brown/none/white |
None/none/red/none |
| Strp 23 |
GAA/SMSA/PGYEA/TA |
Moderate/good/good/good |
Gray/red/red/white |
Gray/red/red/white |
Brown/gray/white/red |
| Strp 24 |
GAA/SMSA/PGYEA/TA |
Moderate/good/good/good |
Gray/white/white/white |
Brown/brown/none/none |
Gray/red/red/white |
| Strp 25 |
GAA/SMSA/PGYEA/TA |
Good/good/moderate/good |
Brown/white/white/none |
Gray/red/red/white |
Brown/white/white/none |
| GAA: Glycerol Asparagine Agar; SMSA: Glycerol Asparagine Agar Medium; PGYEA: Peptone Glycerol Yeast Extract Agar; TA: Throsine Agar |
Table 1: Morphological characteristic of 25 Streptomyces spp. Isolated from agricultural soil region in Baghdad city.
Morphological examination of these isolates indicates that they belong to the genus Streptomyces. The results showed that Streptomyces produced branched substrate and aerial mycelium which upon maturation, they were differentiated into spiral spore chains. The mycelial growth as well as development of spiral spore chains was studied microscopically under a light microscope. Cultural characteristics were determined in glycerol asparagine agar, peptone glycerol yeast extract agar medium Starch mineral salt agar medium, glycerol asparagine agar medium and tyrosine agar. The addition of CaCO3 and heat treatment led to raising the value of hydrogen power which limits the growth of most fungi and increase in growth of actinomycetes.
The result also showed variation in areal mycelium when cultured in different media, it showed that white color is more prevalent between isolates, the reason of change in morphological characteristic of isolates is related to enlargement of nuclear material, it was reached to 10.5 × 10 their DNA has a high GC content, it was reached to 78,69%, so that, it is difficult for classifiers to taxonomy to species. Our result is concordant with saadoun, et al. Also with Al-Obaidi, Antonieta, et al. isolated 71 Streptomyces spp. From soil samples collected at different places of Venezuela the morphological and biochemical characteristics of the isolates indicated that belonged to this species.
The biochemical test showed that, 13 out of 25 isolates in the gray series were able to use all of the tested carbon sources, while in the rest of the series this property was very variable, the isolates grouped in the white series being less efficient in their capacity of utilization of the same carbohydrates. Very few isolates (7 out of 25) were able to use cellulose as carbon source. (12 out of 25) isolates could degrade urea. Regarding the utilization of caseine, tyrosine and xanthine, variable results were also found. Few isolates were unable to degrade tyrosine (2 out of 25) and xanthine (5 out 25), but only 20 isolates out of 25 could degrade casein.
To more confirm that these isolates were belonged to streptomycin all 25 isolates were identified using PCR reactions using primer pair, the result showed a single of about 1500 bp band in all tested isolates. Our result are similar with Hadi, et al., from 140 isolates collected northwest of Iran, 12 selected Streptomyces isolates which exhibited high antibacterial activity against pathogenic bacteria were subjected to PCR reaction for identification via 16S rDNA gene, they obtained a specific band of about 1500 bp (Figure 1).
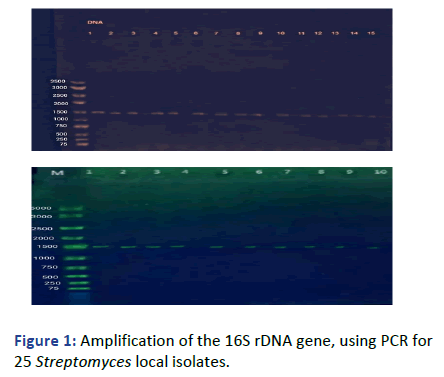
Figure 1: Amplification of the 16S rDNA gene, using PCR for 25 Streptomyces local isolates.
Antimicrobial Activity
In this study, only 10 streptomyces isolates showed antimicrobial activity against colonies of Escherichia coli, Staphylococcus aureus, Bacillus subtilis, Pseudomonas aeruginosa, it was observed isolates have varied antimicrobial activity of Streptomyces secondary metabolites. Our result are concordant with Latif, et al. is five strain of Streptomyces whose namely S,N,W, E and C. Isolated from rizospher soil cultivated with palm, Al-madina city, Saudi arabia were that are indicated produce antibiotic (Table 2).
| Isolates |
E. coli |
S. aureus |
B. cubtilis |
P. aeruginosa |
| Strep 1 |
5 |
11 |
10 |
2 |
| Strep 10 |
5 |
12 |
15 |
8 |
| Strep 11 |
8 |
30 |
10 |
10 |
| Strep 15 |
6 |
13 |
30 |
9 |
| Strep 16 |
3 |
22 |
20 |
2 |
| Strep 18 |
4 |
10 |
10 |
5 |
| Strep 19 |
8 |
12 |
10 |
4 |
| Strep 21 |
5 |
25 |
12 |
3 |
| Strep 23 |
2 |
5 |
11 |
4 |
| Strep 24 |
5 |
11 |
10 |
5 |
Table 2: Comparison of antimicrobial activity of local Streptomyces spp. against standard clinical bacteria.
It is clear from above table. It is seems that S. aureus and B. subtilis have more inhibition zone than E. coli and P. aeruginosa which supports the findings of Scherer and Gerhardt who stated that gram positive bacteria are more sensitive to metabolites produced by Streptomyces. Antibiotics are the most vital bio active secondary metabolites for the treatment of infectious diseases. Since last two decades, Multi Drug Resistance (MDR) increased due to absence of new antibiotic that is a basic challenge for effective treatment of infectious diseases. Due to increased burden of multidrug resistance, there has been increasing interest of researcher for searching of novel bio active secondary metabolites to overcome multidrug resistance of pathogens. Actinomycetes have potential to produce novel bio active secondary metabolites to cure infectious disease. For this purpose different soil samples were taken from different spots for isolation of actinomycetes. Previous research studies showed that novel actinomycetes are most prominently found in soil so soil samples were an important activity for isolation of antibiotic producing soil actinomycetes.
According to previous studies, in inoculated plates, clear zones around the wells are an indication of antimicrobial activity of actinomycetes secondary metabolites against test organisms. Gurung, et al., reported that 0 mm-18 mm inhibition zone of actinomycetes secondary metabolites against test organisms. From present study a range of recorded inhibition zone of actinomycetes secondary metabolites was 2 mm-30 mm which is higher than reported by Gurung, et al., [16-20].
Influence of some cultivation factors on the production of antibiotic: In this study, we examined the impact of various carbon and nitrogen molecules on the generation of antimicrobial by a number of isolated strep 15 (Table 3).
| Carbohydrate |
Biomass g/L |
E. coli |
S. aureus |
B. cubtilis |
P. aeruginosa |
| Glucose |
16.36 |
- |
15 |
9 |
2 |
| Maltose |
15.26 |
4 |
14 |
10 |
5 |
| Sucrose |
6.2 |
1.5 |
- |
- |
- |
| Lactose |
8.26 |
- |
2 |
- |
- |
| Starch |
5.04 |
- |
|
3 |
2 |
Table 3: Effect of carbohydrates on strep15's ability to produce antibiotics.
A number of carbohydrate were investigated for their influence on growth one selected Streptomyces and on its antibiotic production. The results of the experiment demonstrate that glucose is a great carbon source for the synthesis of antibiotics, leading to enhanced antimicrobial production. The inhibition zone shows that S. regions produced 15 mm diameter and the inhibition zone. Our results are concordant with Al-obaidi who isolated Streptomyces spp. From rizosphere of agricultural region in Mosul city when added glucose to basal medium, she used different concentration of glucose to test the influence on growth and production of antibacterial, she obtained higher antibiotic production when adding glucose at concentration 15 gram per litter and inhibition zone 20 mm against S. aureus. Our results showed that when maltose added to media was reached to 14 mm against S. aureus. While when sucrose and starch used as a source carbon source for production of antibiotic give poor production. It's probable that these carbon sources are quickly used for the synthesis of cellular components, leaving little carbon and energy for the production of antibiotics. Our findings are in agreement with those of Pandey, et al. who investigated a variety of carbon and nitrogen compounds for their impact on Streptomyces kananmyceticus M27's synthesis of antibacterial antibiotics. Although maltose, sucrose, and soluble starch produced only moderate yields, dextrose was shown to be the most suited carbon source. For the synthesis of antibiotics, (NH4)H2PO4 and yeast extract were suitable nitrogen sources (Table 4).
| Con. of (NH4)2SO4 |
Biomass |
E. coli |
S. aureus |
B. subtils |
P. aurgenosa |
| 0.05 |
14.2 |
10 |
12 |
10 |
9 |
| 0.1 |
20.6 |
12 |
14 |
13 |
10 |
| 0.15 |
19.6 |
10 |
12 |
12 |
12 |
| 0.2 |
16.8 |
10 |
12 |
12 |
10 |
Table 4: Concentration of ammonium sulfate on growth and antimicrobial production of clinical bacteria.
Different concentration NH4SO4 were added to optimize the optimal concentration of antimicrobial production which added to fermentation medium as nitrogen source, it is clear from table, higher production when added at concentration 0.10 which showed a clear inhibition zone against S. aureus and B. subtilist which reached to 14 mm-13 mm respectively. Nitrogen source play an important source to determine antimicrobial production. Fraid, et al., indicated that higher production of antimicrobial natamycin produced from S. natalensis when nitrogen source at concentration 8 and 2 g/l, better antimicrobial production where 2.5 g/l when ammonium natrite added as nitrogen source. A large number of antibiotics are synthesized with the help of phosphate. But too much inorganic phosphate prevents the synthesis of antibiotics like tetracycline, actinomycin, and candicidin. Results are consistent with those provided by other researchers.
Conclusion
We concluded from our study that isolated microorganism from rizosphere soil of agriculture region in Baghdad city were identified belonged to Streptomyces genus, and these isolates capable to produce antimicrobial substance and this indicates that the soil region are important source foir production new secondery metabolites. Extensive study will be carried out in the future to explore more bioactive compounds from this region. Antibiotics production by these isolates could easily be detected on solid medium. The present study conducted that the number of member of Streptomyces genus in selected soil in Baghdad city/Iraq representing new resource for bioactive production in the biotechnology. Studies are currently being conducted to produce bioactive compounds from actinobacteria through fermentations of different substrates.
References
- Pathalam G, Rajendran HAD, Appadurai DR, Gandhi MR, Michael GP, et al. (2017) Isolation and molecular characterization of actinomycetes with antimicrobial and mosquito larvicidal properties. Int J Basic Appl. 6(2):209-217.
- Rajivgandhi G, Vijayan R, Kannan M, Santhanakrishnan M, Manoharan N (2016) Molecular characterization and antibacterial effect of endophytic actinomycetes Nocardiopsis sp. GRG1 (KT235640) from brown algae against MDR strains of uropathogens. Bioact Mater. 1(2):140-150.
[Crossref] [Google Scholar] [PubMed]
- Adegboye MF, Babalola OO (2016) Isolation and identification of potential antibiotic producing rare actinomycetes from rhizospheric soils. Hum Ecol. 56(2):31-41.
[Crossref] [Google Scholar]
- Khebizi N, Boudjella H, Bijani C, Bouras N, Klenk HP, et al. (2018) Oligomycins A and E, major bioactive secondary metabolites produced by Streptomyces sp. strain HG29 isolated from a Saharan soil. J Med Mycol. 28(1):150-60.
[Crossref] [Google Scholar]
- Xia H, Li X, Li Z, Zhan X, Mao X, et al. (2020) The application of regulatory cascades in Streptomyces: Yield enhancement and metabolite mining. Front Microbiol. 11(1):406-415.
[Crossref] [Google Scholar] [PubMed]
- Hassan SED, Fouda A, Radwan AA, Salem SS, Barghoth MG, et al. (2019) Endophytic actinomycetes Streptomyces spp mediated biosynthesis of copper oxide nanoparticles as a promising tool for biotechnological applications. J Biol Inorg Chem. 24(3):377-393.
[Crossref] [Google Scholar] [PubMed]
- Krzesniak KJ, Mateusiak AR, Guspiel A, Ziemska J, Solecka J (2018) Secondary metabolites of actinomycetes and their antibacterial, antifungal and antiviral properties. Pol J Microbiol. 67(3):259-267.
[Crossref] [Google Scholar] [PubMed]
- Frieri M, Kumar K, Boutin A (2017) Antibiotic resistance. J Infect Public Health. 10(4):369-378.
[Crossref] [Google Scholar] [PubMed]
- Berendonk TU, Manaia CM, Merlin C, Kassinos DF, Cytryn E, et al. (2015) Tackling antibiotic resistance: The environmental framework. Nat Rev Microbiol. 13(5):310-317.
[Crossref] [Google Scholar] [PubMed]
- Takahashi Y, Nakashima T (2018) Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics. 7(2):45-56.
[Crossref] [Google Scholar] [PubMed]
- Lin J, Nishino K, Roberts MC, Tolmasky M, Aminov RI, et al. (2015) Mechanisms of antibiotic resistance. Microbiol Spectr. 6(1):34-42.
[Crossref] [Google Scholar] [PubMed]
- Patel P, Patel G, Mehta P (2020) Extraction and molecular characterization of antimicrobial metabolites from Streptomyces rochei against bacterial leaf blight of cotton caused by Pantoea sp. Asian J Biol Sci. 9(2):159-167.
[Google Scholar]
- Kum E, Ince E (2021) Genome guided investigation of secondary metabolites produced by a potential new strain Streptomyces BA2 isolated from an endemic plant rhizosphere in Turkey. Arch Microbiol. 7(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Zothanpuia AKP, Chandra P, Leo VV, Mishra VK, Kumar B, et al. (2017) Production of potent antimicrobial compounds from Streptomyces cyaneofuscatus associated with fresh water sediment. Front Microbiol. 8(1):1-13.
[Crossref] [Google Scholar] [PubMed]
- Ouchari L, Boukeskasse A, Bouizgarne B, Ouhdouch Y (2010) Antimicrobial potential of actinomycetes isolated from the unexplored hot Merzouga desert and their taxonomic diversity. Biology Open. 8(2): 35410.
[Google Scholar] [PubMed]
- Atashpaz S, Khani S, Barzegari A, Barar J, Vahed SZ, et al. (2010) A robust universal method for extraction of genomic DNA from bacterial species. Microbiology. 4:538-542.
[Google Scholar] [PubMed]
- Bizuye A, Moges F, Andualem B (2013) Isolation and screening of antibiotic producing actinomycetes from soils in Gondar town, North West Ethiopia. Asian Pac J Trop Dis. 3(5):375-381.
[Google Scholar]
- Ilic SB, Konstantinovis SS, Todorovic ZB (2005) UV/V15 analysis and antimicrobial activity of Streptomyces isolates. Med Biol. 12:44-46.
[Google Scholar]
- Anansiriwattana W, Tanasupawat S, Amnuoypol S, Suwanborirux K (2006) Identification and antimicrobial activities of actinomycetes from soils in Samed Island, and geldanamycin from strain PC4-3. Thai J Pharm Sci. 30(2006):49-56.
[Google Scholar]
- Amade P, Mallea M, Bouaicha N (1994) Isolation, structural identification and biological activity of two metabolites produced by Penicillium olsoniibainier and Sartory. J Antibiot. 47(2):201-207.
[Crossref] [Google Scholar] [PubMed]
Citation: Najm BY, Khallel SH, Majeed HM (2023) Evaluation of the Antimicrobial Producing Actinomycetes from Regions in
Baghdad City. Biochem Mol Biol J. 9:32.
Copyright: © 2023 Najm BY, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


