Himani Dhingra1*, Manas Kalra1, Leena Mendiratta2, Hena Butta2, Raman Sardana2 and Amita Mahajan1
1Pediatric Oncology Unit, Indraprastha Apollo Hospitals, New Delhi, India
2Department of Microbiology, Indraprastha Apollo Hospitals, New Delhi, India
- *Corresponding Author:
- Himani Dhingra
Pediatric Oncology Unit
Indraprastha Apollo Hospitals, New Delhi, India
Tel: + 07015671278
E-mail: drhimani458@gmail.com
Received Date: October 10, 2019; Accepted Date: December 25, 2019; Published Date: January 03, 2020
Citation: Dhingra H, Kalra M, Mendiratta L, Butta H, Sardana R, et al. (2020) Fighting the Rising Spectre of Tough Bugs in Tough Clinical
Scenario: A Ten Year Experience from a Hemato-Oncology Centre in Tertiary Health Care Facility in North India. J Infec Dis Treat Vol.6 No.1:1.
DOI: 10.36648/2472-1093.6.1.47
Keywords
Blood culture; Anti-microbial resistance; Sepsis; Antibiotic stewardship; Pediatric oncology
Introduction
Increased susceptibility to infections, consequent to immunosuppressive therapy is a major cause of morbidity and mortality in patients with malignancy. The alarming rise in prevalence of Multi drug resistant (MDR) organisms pose a major challenge in achieving optimal outcomes in pediatric oncology patients [1]. With no new class of antibiotics discovered over the last two decades, this fight against superbugs is even more challenging [2]. The appropriate management of febrile neutropenia (FN) mandates periodic review of the epidemiology of blood stream infections (BSI) and the antibiotic sensitivity pattern. This will help formulate individualized empirical antimicrobial policy for every oncology unit and hence rationalize antibiotic use. An antibiotic stewardship programme helps curb the menace of drug resistance. However the impact of such a programme has not been studied widely in developing countries. Through this study we aim to analyze the evolving blood stream flora, its antibiotic resistance pattern and impact of a rigorous antibiotic stewardship programme at our centre.
Materials and Methods
This is a ten year retrospective, observational analysis of data from pediatric hematology and oncology unit of a tertiary care centre in North India. The study cohort included all patients (<18 years of age) admitted in our unit with suspected episodes of septicemia over the period of 2007-2018. Blood culture samples for all these patients were sent either from central venous catheters (PICC line/chemoport) or peripheral vein before starting the antimicrobial therapy. Adequate volume of blood was withdrawn with all aseptic precautions as per the recommendations. Bacterial isolates were identified both manually and using an automated system. In-vitro antimicrobial susceptibility testing was done by automated Vitek-2 system and interpretation was done as per Clinical and Laboratory Standards Institute (CLSI) guidelines. Blood culture isolates, resistance patterns and clinical outcome of corresponding infection episodes were studied over two time periods-TP1 (2007- 2013) and TP2 (2014-2018).
The choice of first line empiric antibiotic for FN in our unit was monotherapy with cefoperazone/sulbactum. Repeat blood cultures were sent for neutropenic patients with persistent fever after 72 hours or earlier if there was any clinical deterioration while on antimicrobial therapy. Antibiotic escalation/ de-escalation approach was followed based on physician advice or sensitivity report. Wherever indicated, Carbapenems were used as the second line agents. Teicoplanin or Vancomycin was added in patients with suspected catheter related sepsis, soft tissue infection or severe mucositis. Prophylactic antibiotics were not used as a unit policy.
Results
A total of 1423 blood culture samples were sent over ten year study period, of which 285 were positive. The cumulative culture positivity rate was 20%. There was no significant difference in culture positivity rate among TP1 (22.7%) and TP2 (18.1%).
Isolates
TP1 showed predominant growth of Gram negative bacteria (GNB-48.5%) and TP2 showed isolates that were mainly Gram positive cocci (GPC-50.3%) reflecting the increased use of central lines. The spectrum of blood stream isolates in TP1 and TP2 are depicted in Table 1. Coagulase negative staphylococcus (CoNS) was the commonest GPC isolated. Enterobacteriaceae accounted for 48.9% of the GNB in TP2.
Table 1: Comparative analysis of blood stream isolates over time point 1 (TP1) and time point 2 (TP2).
| Isolates |
TP1 |
TP2 |
| Gram Negative Bacteria |
48.50% |
40.30% |
| Enterobacteriaceae |
57.80% |
48.90% |
| Non-fermenting bacteria |
40.60% |
48.90% |
| Aeromonas sp. |
Single isolate |
Single isolate |
| Gram Positive Cocci |
41% |
50.30% |
| Coagulase negative Staphylococcus |
65.00% |
63.80% |
| Staphylococcus aureus |
11.10% |
9.72% |
| Non Enterococcus Streptococcus |
9.50% |
9.72% |
| Enterococcus sp. |
NIL |
4.16% |
| Streptococcus pneumoniae |
1.50% |
4.16% |
| Fungi |
5.90% |
7.90% |
| Candida albicans |
28.50% |
33.30% |
| Non albicans Candida |
71.40% |
66.70% |
The individual GNB isolated in decreasing order of frequency in TP1 and TP2 were Escherichia coli (35%, 19.6%), Klebsiella sp. (12.3%, 16.3%) and P. aeruginosa (6%, 14.7%). The non-albicans Candida was the most commonly isolated yeast both in TP1 and TP2. Changing flora in TP2 with emerging virulent bacterial pathogens like Raoutella, Ralstonia, Citrobacter and fungal pathogens like Candida auris was observed. Non albicans Candida as the most commonly isolated yeast in both TP1 and TP2.
Resistance pattern in GPC
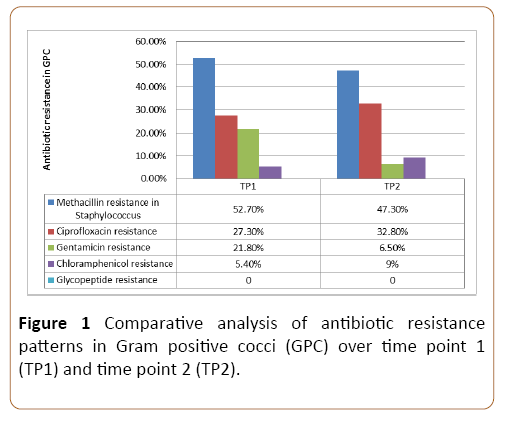
The methicillin resistance in GPC continued to be high over TP1 (52.7%) and TP2 (47.3%). Majority of them were sensitive to chloramphenicol and gentamicin. There was no in vitro resistance to glycopeptides antibiotics. The comparative resistance pattern of GPC over TP1 and TP2 is shown in Figure 1.
Figure 1: Comparative analysis of antibiotic resistance patterns in Gram positive cocci (GPC) over time point 1 (TP1) and time point 2 (TP2).
Resistance pattern in GNB
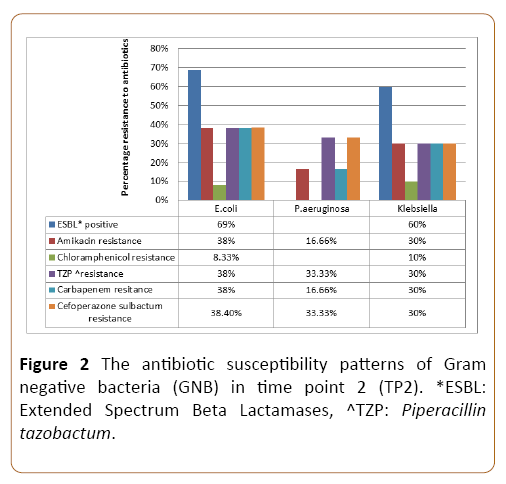
In TP2, the overall rate of extended spectrum beta lactamase (ESBL) production was highest among E. coli (69%) followed by Enterobacter (66.6%) and Klebsiella (60%) respectively. GNB showed good susceptibility to Amikacin with Acinetobacter being most susceptible (85.8%) and E. coli being the least (62%). E. coli (38%) and Klebsiella (30%) showed significantly high in vitro resistance to Carbapenems. The three most common GNB isolates showed around 35-40% resistance to Piperacillin and Tazobactam. Almost 72% of GNB were susceptible to cefoperazone sulbactum with maximum in vitro resistance seen in E. coli. There was no resistance identified to Colistin over this 10 year study period. The antibiotic susceptibility of commonly isolated GNB in TP2 is shown in Figure 2.
Figure 2: The antibiotic susceptibility patterns of Gram negative bacteria (GNB) in time point 2 (TP2). *ESBL: Extended Spectrum Beta Lactamases, ^TZP: Piperacillin tazobactum.
Comparative analysis of resistance patterns over TP1 and TP2
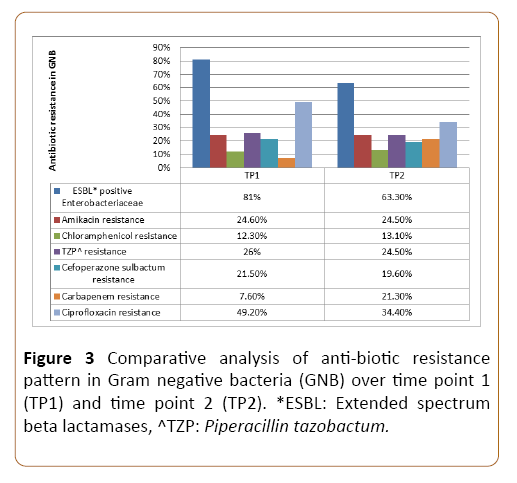
Of all the Enterobacteriaceae 81.25% in TP1 and 63.3% in TP2 were ESBL producing isolates. There was emerging Carbapenem resistance observed in TP2 (21.3%) as compared to TP1 (7.6%). Almost 87% of the GNB isolates over TP1 and TP2 were susceptible to chloramphenicol, making it an attractive alternate for MDR bacteria wherever feasible. Fluoroquinolone resistance (71.8%) has consistently been high making it a poor drug of choice for prophylaxis or therapy. Figure 3 highlights the comparative antibiotic resistance pattern of GNB over TP1 and TP2.
Figure 3: Comparative analysis of anti-biotic resistance pattern in Gram negative bacteria (GNB) over time point 1 (TP1) and time point 2 (TP2). *ESBL: Extended spectrum beta lactamases, ^TZP: Piperacillin tazobactum.
Outcomes
Mortality rates in children with sepsis who had a positive blood culture remained the same across TP1 and TP2 (2.2 vs. 2.6%).
Discussion
Mortality due to infection is one of the most common avoidable causes of deaths in oncology. Children with malignancy represent a unique population that is extremely vulnerable to infections because of alterations in host defense mechanisms. This may be either related to the underlying illness or to the side effects of the intensive treatment with immuno-suppressive drugs leading to neutropenia, lymphopenia or mucositis. Frequent presence of the central lines complicates this issue further. Neutropenic fever (FN) is a life threatening medical emergency that mandates appropriate and timely intervention. Periodic review of the epidemiology of BSI and the antibiotic sensitivity pattern is imperative for management of FN. This helps formulate individualized empirical antimicrobial policy for every oncology unit and hence rationalize antibiotic use.
In our study the culture positivity rate was 20% and has been the same over the ten year study period. This is comparatively much higher than the rates reported from other oncology centers (<10%) in India [3,4] but is similar to the data from the developed countries (11-38%) [5,6]. This difference could be attributed to the increased use of central venous catheters at our center and adequate aliquots of blood being withdrawn for sampling. In contrast to previous studies from India where GNB was the most common isolate [7,8], our study showed GPC as the predominant isolate in TP2 which also is likely due to the increased use of central venous access and no policy of using prophylactic antibiotics. The GPC: GNB ratio in TP2 in our study is comparable to that reported in European multicenter study by Mikulska et al. [9].
Likewise, Mvalo et al. have also reported maximum BSI in their pediatric oncology unit due to GPC (49.1%) followed by GNB (41.6%) and fungi (9.3%). Amongst GPC the maximally isolated bacteria were CoNS (23.1%) followed by Viridans group of Streptococci (13.3%) and amongst GNB the predominant bacteria were Escherichia coli (11%) and Klebsiella species (10.4%) [10].
Resistant GNB are a major cause of concern in most of the oncology centers. Among Enterobacteriaceae, Klebsiella has been reported to be the most problematic bug showing increased antibiotic resistance [3] and also the major attribute to septic deaths [8], however in our study E. coli showed higher resistance to antibiotics. Radhakrishnan et al. [11] have reported marked resistance to cefoperazone/sulbactum in E. coli and Klebsiella, as opposed to our study where it showed good susceptibility. Similarly higher Aminoglycoside resistance among GNB has been reported from different centers [11] as compared to our study. Markedly increased Carbapenem resistance was observed in TP2 in our study (21.3%) which however is less than previously reported in studies [3,4]. There was no colistin resistance, which has been reported previously from various oncology centres [3,8]. However GNB isolates have recently shown colistin resistance by redefined Colistin MIC (Minimum Inhibitory Concentration) testing criteria. Previous studies [8,12] have even reported extreme drug resistant and pan drug resistant gram negative isolates but this was not seen in our study. Interestingly, almost 87% of the GNB isolates over TP1 and TP2 were susceptible to Chloramphenicol, making it an attractive alternate for MDR bacteria wherever feasible.
Though most of the centres have reported good antibiotic susceptibility among Staphylococcus aureus and other GPC, but the overall rate of methicillin resistant CoNS in our center is much higher than reported previously in similar type of study by Thacker et al. [3]. Most frequently isolated CoNS from BSI include Staphylococcus haemolyticus, S. epidermidis, S. hominis and S. warneri. However, S. haemolyticus per se showed more resistance in comparison to other Staphylococci. In other studies also on BSI by CoNS higher resistance rate to methicillin has been reported [13]. There were no Enterococcus spp. isolates in TP1 and amongst the ones isolated in TP2 none was Vancomycin resistant Enterococcus (VRE). This has been reported previously by Thacker et al. [3] Prabhash et al. [7] and Trehan et al. [14].
Mortality rates in patients with culture positive bacteremia were significantly lower in our study in comparison to previous studies [14]. Also the mortality rates remained same over two time periods in our study. It is due to effective case management based on early accurate diagnostics, timely communication from laboratory, implemented infection control programme (Hand hygiene compliance >90% amongst all staff at any time), a restricted antimicrobial policy and early directed antimicrobials.
Conclusion
Despite rising incidence of MDR bacteria, a rigorous antibiotic stewardship programme based on implemented measures like early diagnosis, restricted antimicrobials, highly effective infection control program, rigorous hand hygiene practices, de-escalation after initial stabilization, environmental cleaning MDR organism protocol and innovative strategies like domiciliary care guidelines have helped maintain good outcomes in pediatric oncology patients with sepsis.
Author Contributions
Conceptualization and Methodology: Himani Dhingra, Manas Kalra and Amita Mahajan.
Resources and formal analysis: Leena Mendiratta and Hena Butta.
Writing original draft preparation: Himani Dhingra.
Writing review and editing: Manas Kalra, Leena Mendiratta and Hena Butta.
Supervision: Amita Mahajan and Raman Sardana.
References
- El‐Mahallawy HA, El‐Wakil M, Moneer MM, Shalaby L (2011) Antibiotic resistance is associated with longer bacteremic episodes and worse outcome in febrile neutropenic children with cancer. Pediatr Blood Cancer 57: 283-288.
- Devasahayam G, Scheld WM, Hoffman PS (2010) Newer anti-bacterial drugs for a new century. Expert Opin Investig Drugs 19: 215-234.
- Thacker N, Pereira N, Banavali SD, Narula G, Vora T, et al. (2014) Epidemiology of blood stream infections in pediatric patients at a tertiary care cancer centre. Indian J Cancer 51: 438.
- Kapoor G, Sachdeva N, Jain S (2014) Epidemiology of bacterial isolates among pediatric cancer patients from a tertiary care oncology center in North India. Indian J Cancer 51: 420.
- Asturias EJ, Corral JE, Quezada J (2010) Evaluation of six risk factors for the development of bacteremia in children with cancer and febrile neutropenia. Curr Oncol 17: 59.
- Duncan C, Chisholm JC, Freeman S, Riley U, Sharland M, et al. (2007) A prospective study of admissions for febrile neutropenia in secondary paediatric units in South East England. Pediatr Blood Cancer 49: 678-681.
- Prabhash K, Medhekar A, Ghadyalpatil N, Noronha V, Biswas S, et al. (2010) Blood stream infections in cancer patients: A single center experience of isolates and sensitivity pattern. Indian J Cancer 47: 184.
- Reddy R, Pathania S, Kapil A, Bakhshi S (2014) Review of spectrum and sensitivity of bacterial bloodstream isolates in children with malignancy: A retrospective analysis from a single center. Indian J Cancer 51: 425.
- Mikulska M, Viscoli C, Orasch C, Livermore DM, Averbuch D, et al. (2014) Aetiology and resistance in bacteraemias among adult and paediatric haematology and cancer patients. J Infect 68: 321-331.
- Mvalo T, Eley B, Bamford C, Stanley C, Chagomerana M, et al. (2018) Bloodstream infections in oncology patients at Red Cross War Memorial Children’s Hospital, Cape Town, from 2012 to 2014. Int J Infect Dis 77: 40-47.
- Radhakrishnan V, Vijaykumar V, Ganesan P, Rajendranath R, Trivadi G, et al. (2014) Bloodstream infections in pediatric patients at cancer institute, Chennai. Indian J Cancer 51: 418.
- Siddaiahgari S, Manikyam A, Kumar KA, Rauthan A, Ayyar R (2014) Spectrum of systemic bacterial infections during febrile neutropenia in pediatric oncology patients in tertiary care pediatric center. Indian J Cancer 51: 403.
- Singh S, Dhawan B, Kapil A, Kabra SK, Suri A, et al. (2016) Coagulase-negative staphylococci causing blood stream infection at an Indian tertiary care hospital: prevalence, antimicrobial resistance and molecular characterization. Indian J Med Microbiol 34: 500.
- Trehan A, Totadri S, Gautam V, Bansal D, Ray P (2014) Invasive bacterial infections in a pediatric oncology unit in a tertiary care center. Indian J Cancer 51: 428.