- (2006) Volume 7, Issue 3
Rachel M Freathy1, Michael N Weedon1, Beverley Shields1, Graham A Hitman2, Mark Walker3, Mark I McCarthy4, Andrew T Hattersley1, Timothy M Frayling1
1Institute of Biomedical and Clinical Science, Peninsula Medical School. Exeter, United Kingdom
2Centre for Diabetes and Metabolic Medicine, Barts and The London Queen Mary's School of Medicine and Dentistry. London United Kingdom.
3School of Clinical Medical Sciences, University of Newcastle upon Tyne Newcastle upon Tyne, United Kingdom
4Oxford Centre for Diabetes, Endocrinology and Metabolism. Headington, Oxford, United Kingdom
Received: 16 January 2006 Accepted: 11 April 2006
Context Vascular endothelial growth factor (VEGF) is important for pancreatic beta cell development and function. Common variation in the VEGF gene is associated with altered serum concentrations of VEGF and with several diseases, but its role in type 2 diabetes is not known. The single nucleotide polymorphisms C-2578A (rs699947), G- 1154A (rs1570360) and G-634C (rs2010963) in the 5'-region of VEGF are associated with altered serum concentrations of the protein. Objective We performed a large case-control and family-based study to test the hypothesis that these variants are associated with type 2 diabetes in a UK Caucasian population. Participants We genotyped 1,969 cases, 1,625 controls and 530 families for the three single nucleotide polymorphisms. Main outcome measures: Allele and genotype frequencies were compared between cases and controls. Family-based analysis was used to test for over- or under-transmission of alleles to affected offspring. Results :Despite good power (80%) to detect odds ratios of approximately 1.2, there were no significant associations between single alleles or genotypes and type 2 diabetes. Odds ratios (and 95% confidence intervals) comparing case and control allele frequencies for single nucleotide polymorphisms C- 2578A, G-1154A and G-634C were 0.97 (0.88-1.07), 0.99 (0.90-1.09) and 0.97 (0.88- 1.08), respectively. Conclusion This is the first large-scale study to examine the association between common functional variation in VEGF and type 2 diabetes risk. We have found no evidence that these three single nucleotide polymorphisms, shown previously to alter VEGF concentrations, are risk factors for type 2 diabetes in a large UK Caucasian case-control and family-based study.
Association; Polymorphism, Single Nucleotide; Vascular Endothelial Growth Factor A
ALS: amyotrophic lateral sclerosis; CEPH: Centre d’Etude du Polymorphisme Humain; ECACC: European Collection of Cell Cultures; FBAT: familybased association test; SNP: single nucleotide polymorphism; TDT: transmission disequilibrium test; VEGF: vascular endothelial growth factor; YT2D: youngonset type 2 diabetes
Vascular endothelial growth factor (VEGF) is a powerful angiogenic and vascular permeability factor [1]. The VEGF gene consists of 8 exons on chromosome 6p21.3 [2]. Human VEGF pre-mRNA undergoes alternative splicing to give rise to five polypeptide isoforms of 121, 145, 165, 189 and 206 amino acids, whilst translation from an alternative initiation codon results in a larger isoform (L-VEGF) of 395 amino acids [3]. Cells producing VEGF include pancreatic islet, smooth muscle, mesangial, kidney and various tumour cells, along with macrophages, keratinocytes and T cells [4]. There is increasing evidence that, as well as playing a vital role in the development and differentiation of the vascular system, VEGF is critical for cell growth in many tissues.
The VEGF protein is important for pancreatic beta cell development and function. Both exogenous VEGF administration and genetic manipulation to increase endogenous VEGF production promote pancreatic islet revascularisation and prolong islet survival and viability after transplantation in mice [5, 6, 7, 8, 9]. Genetic manipulation in mice has confirmed the key role of VEGF in pancreatic survival. Firstly, there is an increase in islet size and vascularisation in transgenic mice over-expressing Vegfa [10]. Secondly, murine studies of beta cell-specific Vegfa inactivation show lower islet vascularisation, a disordered endothelial structure, and reduced plasma insulin levels following glucose administration [11] or the development of diabetes given a high fat diet to induce insulin resistance, despite a partial compensatory increase in islet mass [12].
Common variation in VEGF is associated with several diseases. Three single nucleotide polymorphisms (SNPs: C-2578A, G-1154A and G-634C) in the VEGF promoter and 5'- UTR are reproducibly associated with amyotrophic lateral sclerosis (ALS). Metaanalysis of four European populations showed that genotypes -2578A/A, -1154A/A and - 634G/G are associated with increased risk of ALS (recessive model; odds ratios (ORs): 1.4 (95% CI: 1.1-1.8), 1.5 (95% CI: 1.1-2.0) and 1.2 (95% CI: 1.0-1.5), respectively) [13]. There is no obvious link between ALS and diabetes but these results suggest that the three SNPs are associated with altered VEGF function. These results are consistent with the observation that ALS patients have low serum VEGF concentrations [13]. There is some evidence of association between VEGF and other diseases, including diabetic retinopathy [14, 15, 16], but these results require further replication due to small sample sizes. There have been no association studies between VEGF and type 2 diabetes risk with a sufficiently large sample size to detect odds ratios of about 1.2.
There is strong evidence that SNPs C-2578A, G-1154A, and G-634C are associated with altered plasma VEGF concentrations and altered VEGF gene expression. Healthy subjects who are either homozygous for the - 1154A [13] allele or who carry at least one copy of the -634G allele [14] have significantly reduced plasma VEGF concentrations. In addition, the AAG and AGG haplotypes of these SNPs are associated with lower plasma VEGF concentrations in two separate Caucasian populations [13]. All three variants are also associated with VEGF production from stimulated peripheral blood mononuclear cells from healthy individuals [17, 18]. Consistent with the in vivo evidence, a luciferase reporter assay confirmed that the - 1154A allele reduces transcription by 25%, while -634G acts post-transcriptionally, reducing production of the L-VEGF isoform by 20% [13]. Additional in vitro work in the same study revealed that the AAG and AGG haplotypes reduce transcription of luciferase by 41% and 30%, respectively.
The evidence that VEGF contributes to the correct functioning of beta cells makes it an important candidate gene for type 2 diabetes susceptibility. We hypothesised that SNPs C- 2578A (rs699947), G-1154A (rs1570360), and G-634C (rs2010963) would be associated with type 2 diabetes in the UK Caucasian population. Here we present the results of a large study examining the role of this common functional VEGF variation in type 2 diabetes.
Subjects for Case/Control Analysis
The clinical characteristics of case and control subjects are shown in Table 1, arranged by sub-group. Case subjects were unrelated UK Caucasians with type 2 diabetes. Cases were included either if they had type 2 diabetes, as defined by World Health Organisation criteria, or if they were being treated with medication for diabetes. Clinical criteria and/or genetic testing were used to exclude known subtypes of diabetes such as maturityonset diabetes of the young or mitochondrial inherited diabetes and deafness. Cases were recruited from three sources, as described previously [19, 20]. These included subjects with young-onset type 2 diabetes (YT2D; age at diagnosis 18-45 years; n=284); probands from type 2 diabetic sibships from the Diabetes UK Warren 2 repository (n=545) [21, 22]; and a more recent collection of subjects with type 2 diabetes from the Diabetes UK Warren 2 repository (age at diagnosis 35-65 years; not selected for having a family history of diabetes; n=1,140). All affected subjects were negative for GAD autoantibodies.
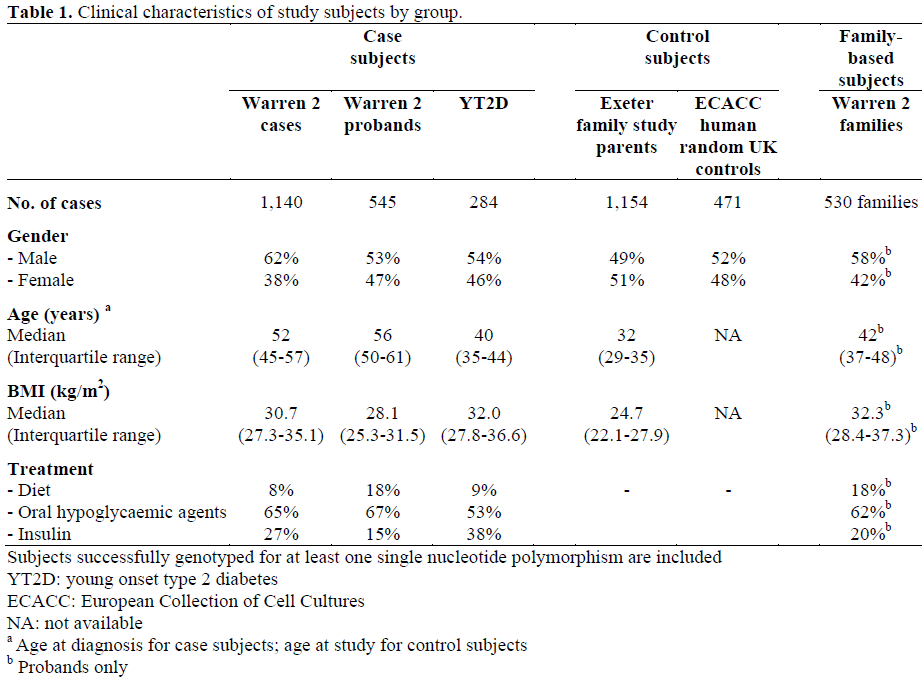
Control subjects were unrelated UK Caucasians recruited from two sources: parents from a consecutive birth study (Exeter Family Study of Childhood Health [23]; n=1,154) with normal (less than 6.0 mmol/L) fasting glucose and/or normal HbA1C (less than 6%, Diabetes Control and Complications Trial-corrected) [22] and a nationally recruited control sample from the European Collection of Cell Cultures (ECACC) (n=471).
Subjects for Family-Based Analysis
Families were recruited as part of a Warren 2 cohort from across the UK. The following criteria were used for inclusion: an affected proband with both parents or with one parent and at least one unaffected sibling (89% of such families included more than two unaffected siblings). The characteristics of some of these families have been described previously [24]. The clinical characteristics of the affected probands are shown in Table 1. Using the same case-control and family-based samples, associations have recently been shown between the KCNJ11, K23 allele [25], the HNF4A P2 promoter haplotype [19] and the PPARG P12 allele [26] and type 2 diabetes with odds ratios consistent with other large type 2 diabetes case-control studies and meta-analyses of multiple studies
Genotyping and Quality Control
Genotyping was carried out by KBiosciences (Herts, UK) using its own novel system of competitive allele specific PCR (KASPar). Details of assay design are available from https://www.kbioscience.co.uk. The genotypeing was performed in 1,536-well plates containing cases, controls and families. At least one negative control well was included per 96 samples. Nine per cent of the samples included were duplicates.
Genotyping accuracy, as determined from the genotype concordance between duplicate samples, was 99.6%. The mean genotyping assay success rate was 97.2% for case and 97.5% for control samples. There were no Mendelian inheritance errors in the familybased sample.
ETHICS
Informed consent was obtained from all participants. The study was approved by local research ethics committee and the protocol conforms to the ethical guidelines of the World Medical Association Declaration of Helsinki.
Before combining case and control subgroups, we tested for homogeneity of genotype and allele frequencies for each SNP using the chi-square test. Odds ratios and P values were calculated using chi-square tests for our case/control analyses. We used COCAPHASE [27] (available at https://portal. litbio.org/Registered/Option/unphased.html; last accessed September 1st, 2005) to estimate haplotype frequencies and perform tests of haplotype association. This program utilises the expectation-maximisation algorithm to account for haplotype uncertainties. To analyse our family data, we used the family familybased association test (FBAT) program [28, 29] (available at https://www.biostat.harvard. edu/~fbat/default.html; last accessed October 12th, 2005). The transmission disequilibrium test (TDT/sibTDT) method [30] was used to confirm the results for single SNPs. Our sample of 1,969 cases and 1,625 controls gave us 80% power to detect odds ratios for alleles of 1.19. This power calculation is for a twotailed P value less than 0.01, assuming a control allele frequency of 0.30 [13].
To assess the degree of linkage disequilibrium between the SNPs chosen for this study and other variants across VEGF, the Haploview package [31] (available at https://www.broad. mit.edu/mpg/haploview/; last accessed September 20th, 2005) was used to analyse both our own data and genotype data (Centre d’Etude du Polymorphisme Humain (CEPH) trios) from The International HapMap Project (Phase I and II data combined; https://www.hapmap.org/index.html.en; last accessed October 14th, 2005).
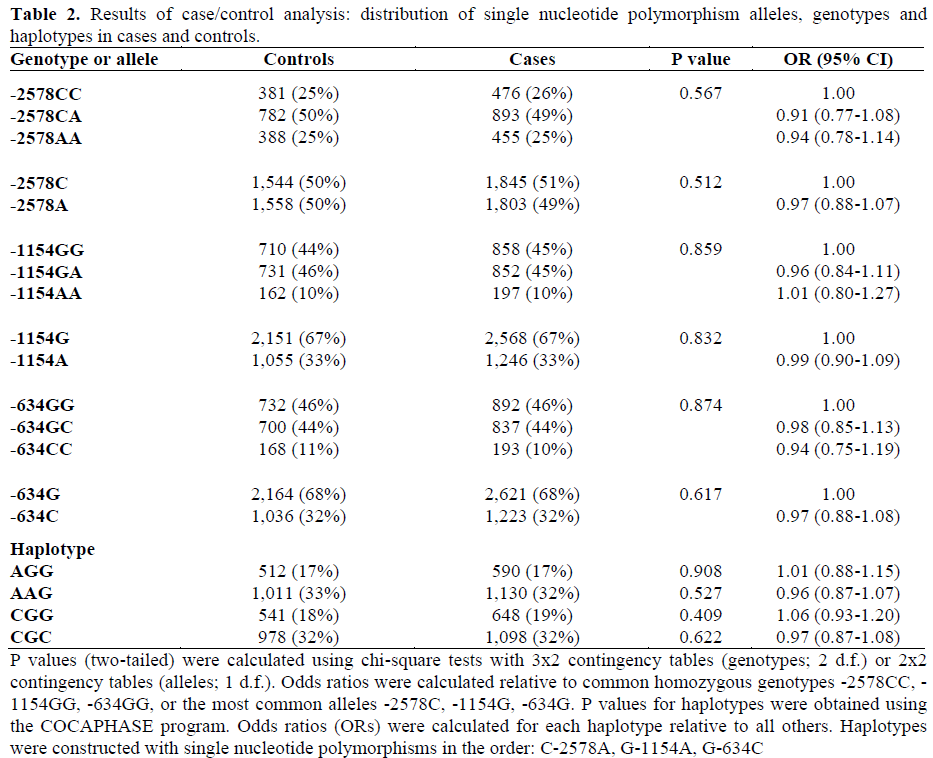
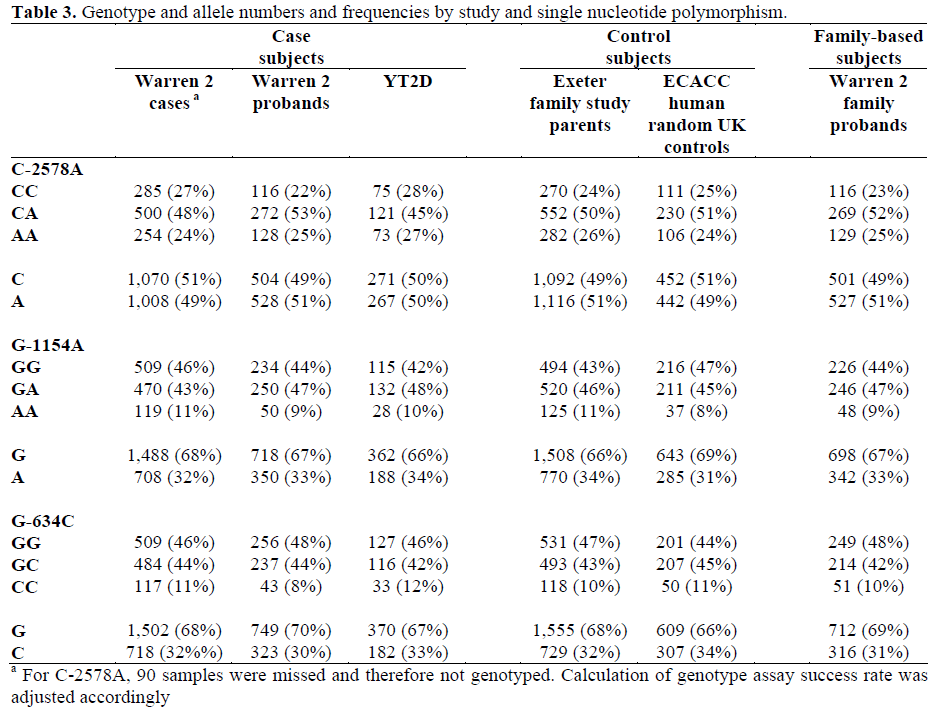
All three SNPs were in Hardy-Weinberg equilibrium in cases, controls and family probands (P>0.102). Hardy-Weinberg equilibrium was also observed within each study subgroup of cases and controls, and the allele and genotype frequencies for each SNP did not differ significantly between these subgroups (P>0.091).
Linkage disequilibrium occurred between SNPs such that only four out of the 8 possible haplotypes were observed. Pairwise r2 values between the three SNPs were all less than 0.5 (maximum r2 equal to 0.484), meaning that they provide non-redundant information. Control allele and haplotype frequencies were similar to those observed previously [13]. Analysis of HapMap genotype data showed that the only SNP in this study for which HapMap genotype data are available (C- 2578A; rs699947) did not capture (at r2 greater than 0.8) any other of the nine genotyped SNPs across the 20 kb region including VEGF. Genotype, allele and haplotype frequencies for cases and controls are displayed in Table 2. Genotype and allele frequencies are shown separately for each subgroup of cases and controls in Table 3. There were no significant associations between single alleles, genotypes or haplotypes and type 2 diabetes. The result of a global test of haplotype association was P=0.792 (haplotypes excluded with frequency less than 0.05).
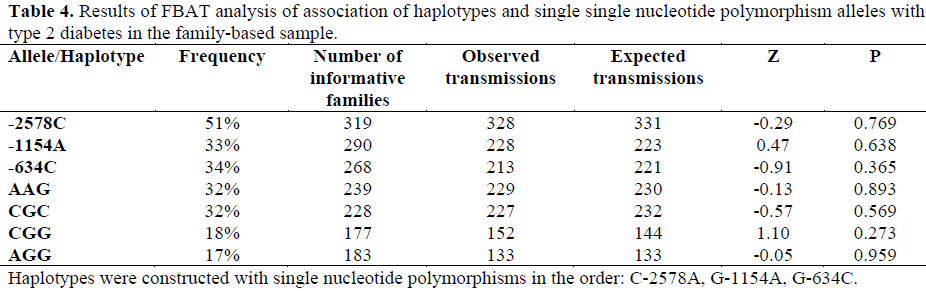
The results of the FBAT analysis are presented in Table 4. There was no significant over- or under-transmission of alleles or haplotypes to the affected offspring. A TDT/sibTDT analysis produced very similar results for single SNPs.
The VEGF gene is a particularly strong type 2 diabetes candidate gene for the following reasons. First, the protein has an important role in pancreatic beta cell function [5, 6, 7, 8, 9, 10, 11, 12]. Second, there is strong evidence that common variation in the gene is functional [13, 14, 18]. We hypothesised that SNPs C-2578A, G-1154A and G-634C, previously associated with altered circulating VEGF levels, would be associated with reduced beta-cell function and hence type 2 diabetes. Our case-control and family-based study results gave no support for this.
Whilst we have not captured all common variation in VEGF, we have tested variants suggested in large cohorts to be functional, and can exclude them from having all but a minor effect (OR less than 1.14, 1.27 and 1.19 for SNPs C-2578A, G-1154A and G-634C, respectively) on type 2 diabetes susceptibility. It is possible that the relatively young age of our control subjects compared to our cases reduced our power to detect an association: some of our controls may yet develop type 2 diabetes. However, this is unlikely to affect more than 5-10% of Caucasian subjects [32]. In addition, most of our case subjects were ascertained for family history or young-onset (or both), which means they are likely to be enriched for genetic effects. The same data set has been sufficiently powerful to detect association for SNPs shown by meta-analysis to be consistently associated with type 2 diabetes [19, 25, 26]. Serum VEGF concentrations were not available for our subjects, so our study is based on the assumption that the SNPs affect VEGF production. However, there is robust evidence for this [13], which is supported by independent work from two other studies [14, 18]. Finally, we have not assessed the role of these polymorphisms in relevant continuous traits, such as measures of insulin secretion. We would expect the potential relationship to be with reduced insulin secretion, due to reduced beta-cell function. Further studies, consisting of large numbers of subjects with oral glucose tolerance test data, will be needed to test this. In conclusion, this is the first large-scale study to examine the association between common functional variation in VEGF and type 2 diabetes. We have examined polymorphisms associated in several studies with altered VEGF concentrations, but found no evidence that they are involved in the genetic susceptibility to type 2 diabetes in our large case-control and family-based study.
RM Freathy holds a Diabetes UK research studentship. AT Hattersley is a Wellcome Trust Research Leave Fellow