Research Article - (2017) Volume 3, Issue 1
Jianying Shen1*,2, Qingsheng Yu3, Xiaoqiang Li1, Rujing Ren1 and Yuqing Tan1
1Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Nanxiaojie, Beijing 100700, China
2Department of Diabetes and Metabolic Disease Research, Beckman Research Institute, City of Hope, Duarte, CA 91010, USA
3Beijing Key Lab for Immune-Mediated Inflammatory Diseases, China-Japan Friendship Hospital, Beijing 100029, People’s Republic of China
*Corresponding Author:
Jianying Shen
Institute of Chinese Materia Medica
China Academy of Chinese Medical Sciences
No. 16, Nanxiaojie, Dongzhimen Nei Ave
Beijing 100700, China
Tel: 861084252805
E-mail: jyshen@icmm.ac.cn
Received Date: January 19, 2017; Accepted Date: February 09, 2017; Published Date: February 15, 2017
Citation: Shen J, Yu Q, Li X, et al. Genome- Wide DNA Methylation Profiling in Diabetogenic and Regulatory T Cells from NOD Mice. J Clin Epigenet. 2017, 3:1. doi:10.21767/2472-1158.100039
Copyright: © 2017 Shen J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
A well-balanced population of pathogenic T (Tpath) and regulatory T (Treg) cells is important for maintaining immune tolerance and preventing autoimmune diseases. However, the molecular mechanisms modulating Tpath or Treg functional differentiation remain largely unclear. Using methylated CpG island recovery assay (MIRA), we have analyzed DNA methylation profiling in one Tpath, two Tregs and one control T cell line (Tctrl). We have identified a total of 2204 different methylation peaks in the four cells and ~1300 peaks in each cell. Analyses of autosomes in Tpath showed relatively lower methylation density than those in Tregs, except for Chromsome 19 that has similar methylation density. Furthermore, 63 hypermethylation and 268 hypomethylation peaks were uniquely found in Tpath, compared to 323 hypermethylation and 30 hypomethylation unique peaks in Tregs. However, the methylation density of Chromosome X in Tpath is significantly higher than that in Tregs. In summary, opposite epigenetic mechanisms are likely involved in methylation of genes on autosomes or Chromosome X in Tpaths and Tregs, thus contributing to the functional differentiation/specification between Tpaths and Tregs.
Keywords
Methylation; Type 1 diabetes; Epigenetics; Autoimmune disease; T cell
Introduction
Type 1 Diabetes (T1D) is an autoimmune disease resulting from selective destruction of insulin-producing pancreatic islet β cells. Previous studies have shown that Monozygotic (MZ) twin pair discordance for childhood-onset T1D is ~ 50% [1,2], implicating that the etiology of T1D involves both genetic and non-genetic components. Many studies have demonstrated the important role of genetic factors in T1D development. For example, it is well established that the major T1D susceptibility locus in human is mapped to the human leukocyte antigen (HLA) class II gene region at chromosome 6p21 [3-6]. Among the most disease-associated HLA alleles, DR3-DQA1*0501-DQB1*0201 (DR3) and DR4-DQA1*0301-DQB1*0302 (DR4) haplotypes account for 30- 50% of T1D incidence in Caucasian population [6]. In addition, other than the HLA genes, genome-wide association studies (GWAS) have also identified more than 40 non-HLA susceptibility loci that may contribute to T1D [7-9]. However, unexpectedly, previous studies also demonstrated that the percentage of T1D cases with high-risk HLA susceptibility genes, such as the DR3/4 genotype, is decreasing as the T1D incidence increases [10,11]. These results suggest the contribution of non-genetic components to the development of T1D.
Emerging evidence has demonstrated that epigenetics is an important non-genetic component in regulating T1D development. Epigenetic changes, including DNA methylation and histone modifications, can be induced by environmental variants including dietary factors and infectious agents and can regulate gene expression levels [12-14]. Therefore, regulation of epigenetic changes is considered an important alternative mechanism to modulate gene expression and functions [15]. An epigenetic study, comparing genome-wide histone H3 lysine 9 dimethylation (H3K9me2) patterns in peripheral blood from T1D patients vs. healthy subjects, has shown that lymphocytes from these two cohorts display a distinct profile of H3K9me2 [16]. In addition, H3K9me2 alteration was detected at the promoter region of CTLA4, one of the T1D susceptibility genes. Studies of other autoimmune disorders, including Systemic Lupus Erythematosus (SLE) and rheumatoid arthritis(RA), further showed that the methylation status of genomic DNA and specific autoimmunity-related genes are altered in patients, leading to autoreactivity [17-19]. A recent study using a systematic analysis of the temporal origins of T1D-associated epigenetic variations, epigenome-wide association study (EWSA), has generated genome-wide DNA methylation profiles of purified CD14+ monocytes from 15 T1D-discordant MZ twin pairs [20]. The results demonstrated the presence of T1D-specific methylation variable positions (T1D-MVPs) in the T1D-affected co-twins. These T1D-MVPs may have arised during the etiological process that leads to overt T1D. Overall, these studies revealed an important role of epigenetic mechanisms in controlling autoimmune diseases, including T1D.
Both Foxp3+CD4+ regulatory T cells (Tregs) and antigen-expanded Foxp3-CD4+ Tregs are important for maintaining immune tolerance and preventing autoimmune diseases, by suppressing pathogenic T cells (Tpaths) [21-28]. These studies suggest that an increased population of autoreactive Tpaths, the presence of Tregs-resistant Tpaths, or a defective population of Tregs may lead to development of diabetes. Therefore, to re-establish a well-balanced population of functionally potent Tregs vs. Tpaths is necessary to help a diabetic patient regain control of their health. To aid in achieving this goal to treat diabetic patients, it is important to understand the molecular mechanisms modulating the homeostatic balance of these functionally varied T cells.
In this study, we performed comparative DNA methylation microarray analyses of Tpaths, Tregs, and a control non-Tpath, non-Treg T cell line (Tctrl). Based on these genome-wide analyses, our studies have found that Tpaths and Tregs have different methylation density distribution in their autosomes and chromosome X. These novel results suggest epigenetic differences may contribute to the functional differentiation/specification of Tpaths vs. Tregs [29-43].
Materials and Methods
Cells CD4+BDC cells(Tpath) [44,45], N79 cells(Tctrl) [46], and two Treg cell lines [47,48], N206 (Treg 1) and NR206 (Treg 2), have been previously described. The features of the cell lines were summarized in Table 1. Ten days after being activated by their activating peptides, the cells were used in the study.
| Cell lines | Features |
|---|---|
| N79 | A CD4+ T cell line, recognizing a mimotope p79 peptide, functions neither as Treg nor Tpath, and is used as functionally neutral control (Tctrl). |
| BDC | CD4+ BDC cells, isolated from the BDC mice, were cultured and activated three times by the p79 peptide in vitro .These BDC cells are able to induce an aggressive form of T1D when transferred to NOD/SCID mice (Tpath). |
| N206 | A GAD peptide p206-specific CD4+ Treg line isolated from nonobese diabetic (NOD) mice, does not require cell contact, but it inhibits Tpath proliferation and prevents T1D in adoptive transfer experiments (Treg1). |
| NR206 | A p206-specific Treg line isolated from nonobese diabetes-resistant (NOR) mice, is dependent on both IFN-γ production and cell contact with target cells (Treg2). |
Table 1 The features of four cells used in the study.
Genomic DNA purification and enrichment of methylated CpG sequences
Genomic DNA was purified using proteinase K digestion. Methylated DNA enrichment was performed as described previously with minor modifications (Rauch and Pfeifer 2010). Briefly, a glutathione-Sepharose CL-4B matrix (Amersham Biosciences, Piscataway) was used to elute puri- fied GST-tagged MBD2b protein with elution buffer, which contained 0.1% Triton X-100, 150 mM NaCl, 50 mM Tris·HCl, pH 8.5 and 20 mM glutathione, at 4°C. Phosphate buffered saline (PBS) was used for dialysis of the eluted GST-MBD2b fraction for 5 h, followed by overnight dialysis in buffer containing 150 mM NaCl, 50 mM Hepes, pH 7.4, 50% (v/v) glycerol and 5 mM 2-mercaptoethanol. GST-tagged MBD2b protein was kept at −20âÂâ€â€Â¦C. Genomic DNA fragments digested by MseI were ligated with linker. Then, after overnight incubation with GST-MBD2b and HisMBD3L1 proteins, MagneGST beads (2.5 μL) (Promega, Madison, USA) which were preblocked by JM110 bacterial DNA, were added into the reaction mixture and incubated at 4âÂâ€â€Â¦C for 45 min. Beads were washed thrice with washing buffer which contained 700 mM NaCl,10 mM Tris·HCl, pH 7.5, 0.1% Triton X-1001 mM EDTA and 3 mM MgCl2. Qiaquick PCR purification kits (Qiagen, Valencia, USA) were used to elute the methylated CpG-enriched fraction. The number of replicated used for MIRA microarray was one.
Definition of CpG methylation peaks: Log2 ratios between MIRA-enriched and input DNA samples were generated using NimbleScan software (Roche NimbleGen) and further processed by Quantile normalization. Probes with log2 ratios greater than 1 (2-fold enriched) were selected as positive. A methylation peak was defined as a region with at least two positive probes covering a minimum length of 250 bp allowing one probe gap, and then mapped relative to known transcripts defined in the UCSC genome browser mm8 RefSeq database. Methylation peaks falling into 1500 bp relative to transcription start sites were defined as ‘promoter’; methylation peaks falling within 1500 bp of RefSeq transcript end sites were defined as ‘downstream’, and those falling within gene bodies (from 1500 bp downstream of transcription start to 1500 bp upstream of the transcript end) were defined as ‘intragenic’ peaks.
Results
Comparative analyses of total methylation peaks in Tpath vs. Tregs.
To gain insights into the potential epigenetic control of the function of CD4+ Tpaths vs. Tregs, we performed comparative genome-wide DNA methylation microarray analyses (methylated CpG island recovery assay, MIRA) of a diabetogenic Tpath (BDC), a non-Tpath/non-Treg control cell Tctrl (N79), and two Treg lines (Treg 1 for N206 and Treg 2 for NR206) (Table 1). The log2 ratios of MIRA signals vs. input were obtained by NimbleGen software with Quantile normalization. The results of probes were considered as positive if their log2 ratios were >2-fold. For our analyses, a methylation peak is defined as having at least 2 positive probes covering a minimum length of 250 bp DNA, allowing no more than a 1-probe gap.
A total of 2204 methylation peaks were positively detected from the CpG methylation array, with 1421, 1493, 1254, and 1159 methylation peaks identified in Treg1, Treg2, Tctrl, and Tpath, respectively. To further analyze the positively detected methylation peaks, we have used the venn diagram (Figure 1) to display the relative number of the common and different methylation peaks in these cells. These results showed that Tpath and Tctrl shared 844 common methylation peaks, while Tpath has 315 and Tctrl has 410 unique methylation peaks. For Treg1 and Treg2, they have 1228 common methylation peaks, while Treg1 has 193 and Treg2 has 265 unique methylation peaks. In addition, there were 635 methylations peaks detected either in Treg1 or Treg2 but not in Tpath or Tctrl, whereas 518 methylation peaks were detected either in Tpath or Tctrl but not in the two Tregs. Our results further showed that ~70% (844/1159=72% for Tpath, and 844/1254=68% for Tctrl) of the methylation peaks were overlapped between Tpath and Tctrl. In comparison, N206 and NR206 Tregs have ~84% (1228/1421=86% for Treg1, and 1228/1493=82% for Treg2) of their methylation peaks overlapped. Altogether, these results indicate that the two Tregs have very similar methylation patterns that are distinct from those found in the Tpath and Tctrl.
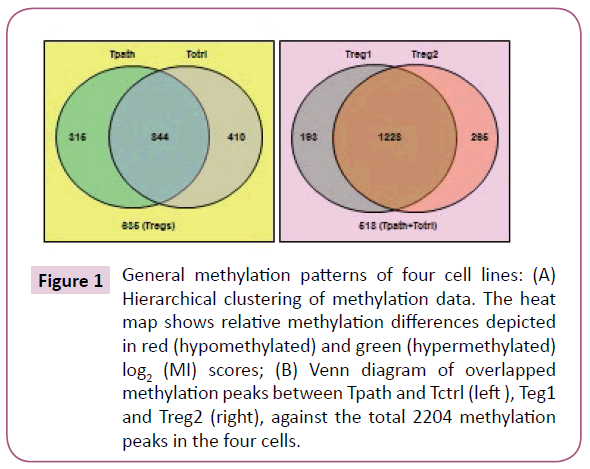
Figure 1: General methylation patterns of four cell lines: (A) Hierarchical clustering of methylation data. The heat map shows relative methylation differences depicted in red (hypomethylated) and green (hypermethylated) log2 (MI) scores; (B) Venn diagram of overlapped methylation peaks between Tpath and Tctrl (left ), Teg1 and Treg2 (right), against the total 2204 methylation peaks in the four cells.
To further analyze the location of various methylation peaks within different regions of certain genes, we adopted the RefSeq gene definition from the University of California at Santa Cruz (UCSC) Genome Browser for mouse (mm8, 22,307 genes) genome. For our analyses, we grouped methylation peaks into four classes (promoter regions, intragenic regions, downstream regions and intergenic regions) based on their distance to RefSeq genes, as defined in Materials and Methods. Based on these definitions, 27-30% of the methylation peaks in each of the four cells is located in the promoter regions, 52-59% in the intragenic regions, and 3% in the downstream regions of a gene. Additionally, 8-15% of methylation peaks are located in the intergenic regions (Figure 2a). Furthermore, there is no difference in the percentage of methylation peaks located in different regions of genes among these cells. Therefore, the number and location of the methylation peaks in different gene regions may not be important for the distinct functions of these T cells. We then analyzed the DNA methylation patterns on each of the 19 autosomes and the X chromosome in the genome of the cells. Our initial analyses showed that the number of methylation peaks along various chromosomes in each type of cells is not proportional to the chromosome size (Figure 2b). These analyses also showed that Tregs have generally higher methylation peaks density (peak number/Mb) than do Tpath and Tctrl on autosomes, except for Chromosome 19 having similar methylation peaks density in these cells. In contrast, unexpectedly, we found that the Chromosome X in Tpath has a significantly higher (~2.5 folds, with 97/166.7 for Tpath, and 39/166.7 for Tregs) methylation peaks density than those in the two Tregs. In addition, the Tpath also has the highest percentage (8%) of their methylation peaks located on Chromosome X (Figure 2c). In comparison, the two Tregs only have 3% and the Tctrl has 4% of their total methylation peaks located on Chromosome X.
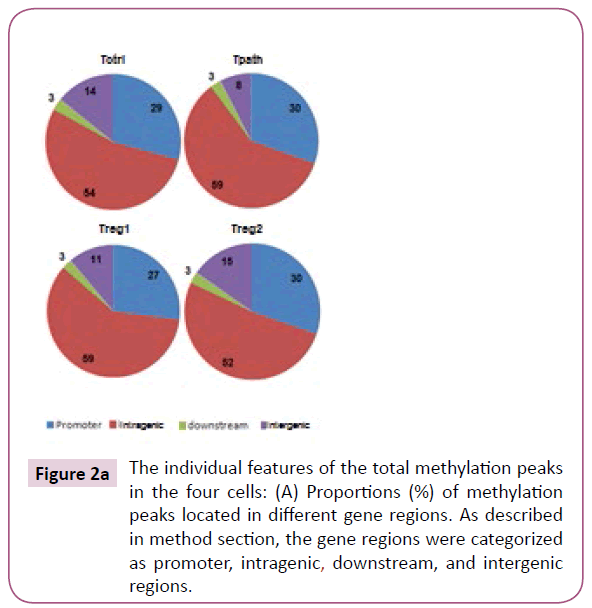
Figure 2a: The individual features of the total methylation peaks in the four cells: (A) Proportions (%) of methylation peaks located in different gene regions. As described in method section, the gene regions were categorized as promoter, intragenic, downstream, and intergenic regions.
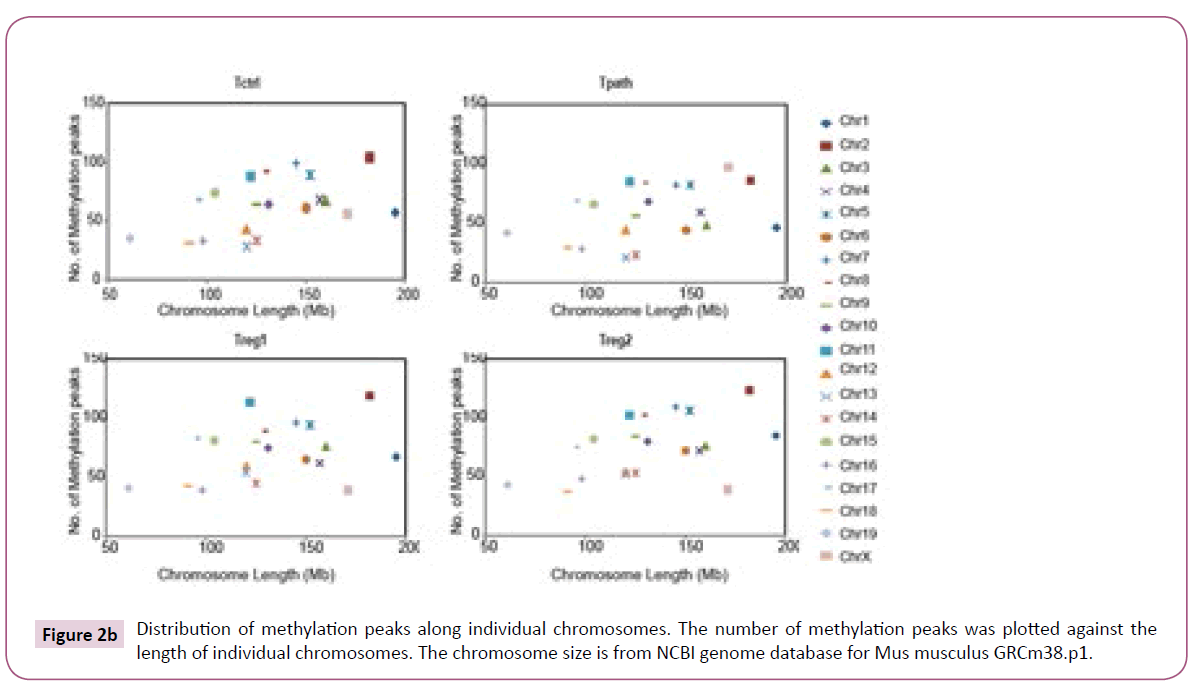
Figure 2b: Distribution of methylation peaks along individual chromosomes. The number of methylation peaks was plotted against the length of individual chromosomes. The chromosome size is from NCBI genome database for Mus musculus GRCm38.p1.
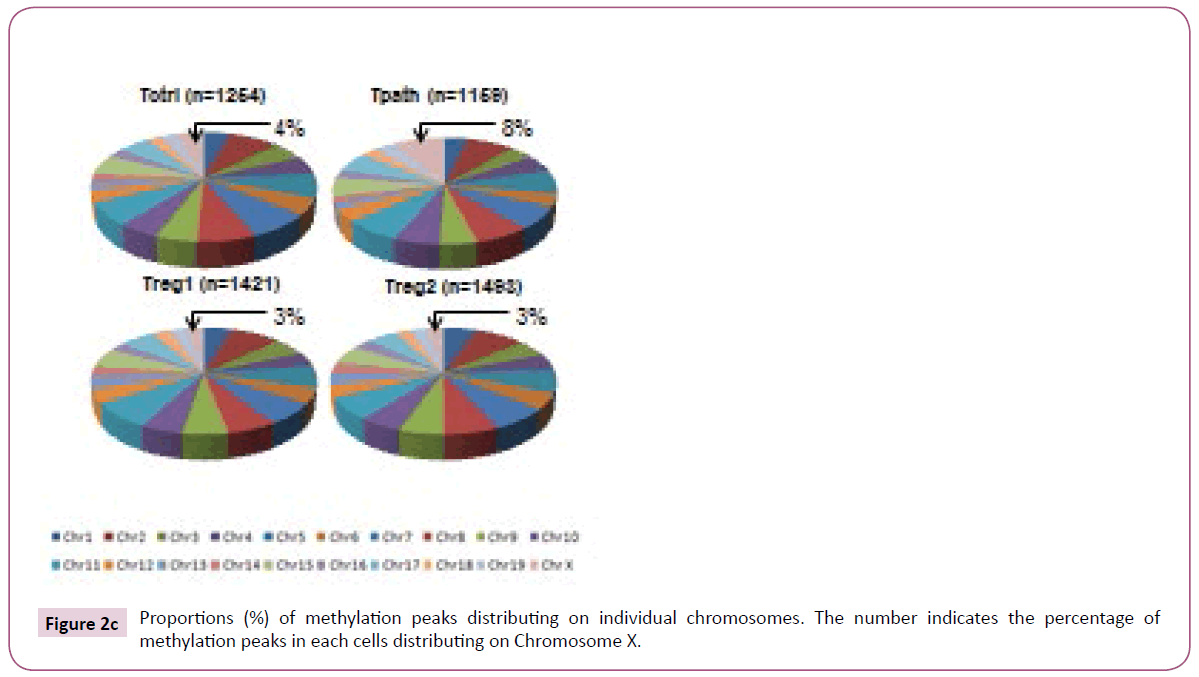
Figure 2c: Proportions (%) of methylation peaks distributing on individual chromosomes. The number indicates the percentage of methylation peaks in each cells distributing on Chromosome X.
These results suggest that higher methylation density on Chromosome X and lower on autosomes (except for Chromosome 19) may be important for the functional differentiation and/ or maturation of Tpath. On the other hand, higher methylation density on autosomes (except for Chromosome 19) and lower on Chromosome X may be important for Tregs.
Comparative analyses of hypermethylation and hypomethylation peaks specific for Tpath or Tregs.
Other than analyses of the total methylation peaks, we further analyzed and compared the relative number and location of hypermethylation or hypomethylation peaks specifically detected in the Tpath or Tregs. Methylation peaks detected only in Tpath or both Tregs are defined as either Tpath-specific or Tregs-specific hypermethylation peaks. On the other hand, methylation peaks detected in the other cells but not in either Tpath or Tregs were defined as Tpath-specific or Tregs-specific hypomethylation peaks, respectively.
A total of 63 hypermethylation and 268 hypomethylation peaks were specifically identified in the Tpath. In comparison, 323 hypermethylation and 30 hypomethylation peaks were specifically identified in the Tregs (Figure 3). Further analyses showed that a vast majority (65%) of Tpath-specific hypermethylation peaks are located mainly in the promoter region, with only 24% in the intragenic region (Figure 3). However, less than 50% of Tregs-specific hypermethylation peaks are located in either the promoter (45%) or the intrageic region (31%). On the other hand, opposite to the hypermethylation peaks, most Tpath-specific hypomethylation peaks are located in either the promoter (36%) or the intragenic region (40%). Whereas, a majority (64%) of Tregs-specific hypomethylation peaks is located mainly in the intragenic region. These analyses also showed that, a significantly fewer Tpath-specific hypermethylation peaks (4%) than that of Tregs-specific hypermethylation peaks (21%) are located in the intergenic region (Figure 3). In contrast, more Tpath-specific hypomethylation peaks (20%) than that of Tregs-specific hypomethylation peaks (10%) are located in the intergenic region. Only a small percentage (3-7%) of cell-specific hypermethylation or hypomethylation peaks are located in the downstream region in these cells. We then analysed the Tpathspecific hypermethylation vs. Tpath-specific hypomethylation peaks on various chromosomes in the genome (Figure 3). Among the 63 identified Tpath-specific hypermethylation peaks,40 peaks (63.5%) are on Chromosome X. Unlike that of Tpath, nearly all of the 323 identified Tregs-specific hypermethylation peaks are on the autosomes. Only one methylation peak is detected on the Chromosome X (0.31%). However, a significantly greater percentage of Tregs-specific hypomethylation peaks (7 out of 30, or 23.3%) are on the Chromosome X, whereas fewer such peaks (23 of the 30, or 76.7%, with a range of 0-4/chromosome) are on autosomes.
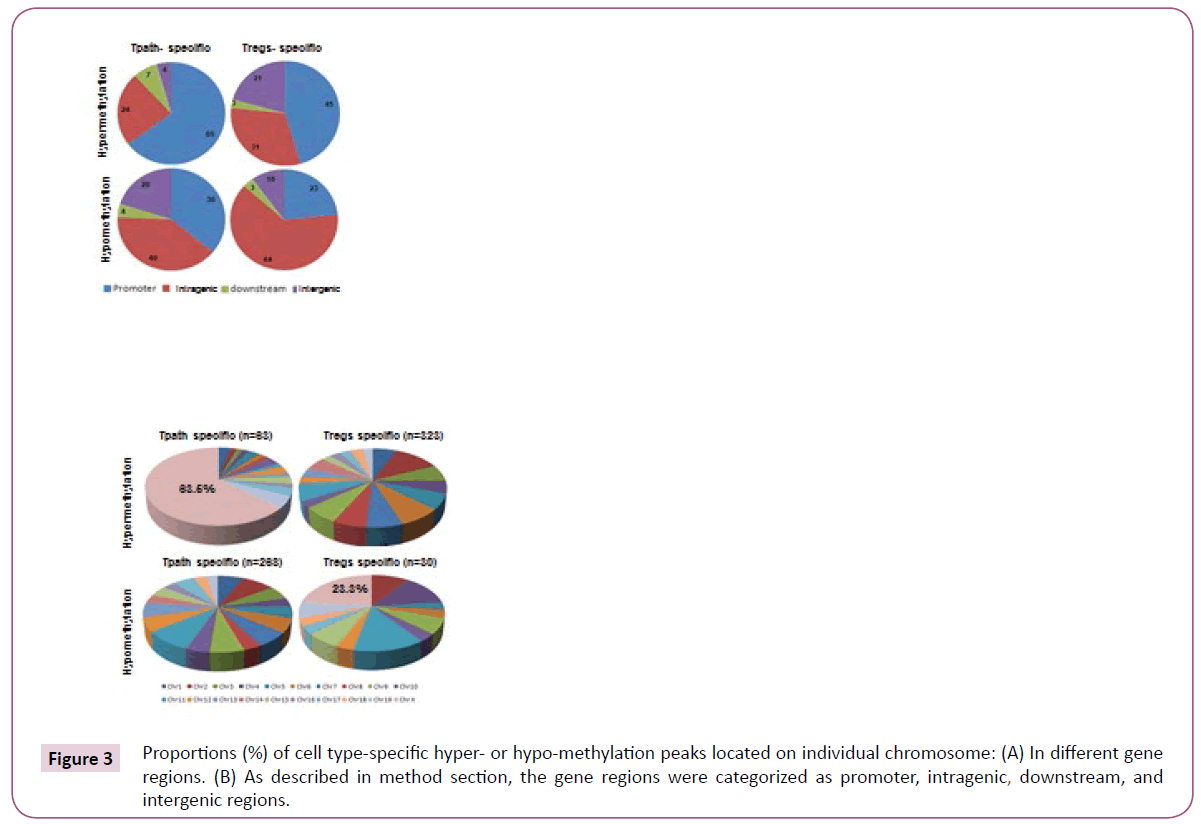
Figure 3: Proportions (%) of cell type-specific hyper- or hypo-methylation peaks located on individual chromosome: (A) In different gene regions. (B) As described in method section, the gene regions were categorized as promoter, intragenic, downstream, and intergenic regions.
Overall, these results may support the conclusion that the dramatic distinct methylation patterns of Chromosome X and autosomes in Tregs vs. Tpaths may play an important role in regulating functional differentiation and/or maturation of these T cells.
Discussion
In this study we performed DNA methylation microarray of one Tpath, two Tregs and one Tctrl in a genome-wide manner. One important discovery is that comparing to Tregs, Tpath shows relatively low methylation density on autosomes, while relatively high methylation density on chromosome X. The discovery can decompose to two aspects: 1) Global DNA methylation of Tpath is lower, especially on autosomes, than that of Tregs. 2) DNA methylation of Tpath on Chromosome X is higher than that of Tregs. The aberrance of DNA methylation in T cells has been linked to pathogenesis of many autoimmune diseases, including SLE [17,29-32], RA [30] and T1D [33]. Thereinto, DNA methylation changes in SLE have been well studied. Global DNA hypomethyation in CD4+ T cells has been detected in SLE. Hypomethylation of specific genes overexpressed in lupus T cells such as ITGAL (CD11a), CD40LG (CD40L), TNFSF7 (CD70), KIR2DL4, IFNGR2 and PRF1 (perforin) is believed to contribute to the development of autoreactivity and overstimulation of autoantibodies [34,35]. Many gene promoters are also found hypomethylated in other autoimmune disorders, such as MET in RA [36], PAD2 in MS [37], and p16, SHP1, p15, and p21 in psoriasis [38-40]. The overexpression of two methyl CpG –binding domain (MBD) proteins, MBD2 and MBD4, may assoiated with the DNA hypomethylation in SLE CD4+ T cells [41]. A genome-wide DNA methylation study with CD4+ T cells from SLE patients revealed 236 hypomethylated and 105 hypermethylated CG sites, and genes involved in autoimmunity, such as MMP9 and PDGFRA were found hypomethylated, while genes involved in DNA methylation were hypermethylated [42]. The DNA methylation studies on T1D are not as much. In a streptozotocin-induced T1D rat model, diabetes induced functional methyl deficiency which resulted in the hypomethylation of NDA in a tissue–specific fashion [43]. A genome-wide DNA methylation study on monocytes from T1D-discodant MZ twin pairs has identified 132 T1D-associated methylation variable positions (T1D-MVPs), which precede disease diagnosis [20]. In our study, we systemically compared the DNA methylations in four cell lines with different functions in NOD mice, a T1D animal model, and found lower methylation in Tpath and higher methylation in Tregs in global genome, and the difference was signified on individual autosomes, except chromosome 19. The difference of DNA methylation between Treg and Tpath may contribute the function difference between these two cells, while the detailed mechanisms need further investigation.
Acknowledgements
The authors thank Wen-Hui Lee for preparing the cell samples in the study and thank Dr. Xiwei Wu for the help with data analysis.