Keywords
Hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX); 16S rRNA; XplA; XplB
Introduction
The explosive hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) is commonly applied to both military and industrial applications. Nowadays, RDX is considering a powerful environmental pollutant as a results of accumulation mainly from weapons manufactures and trainings [1,2]. Also, RDX was listed as possible human carcinogen by U.S. Environmental Protection Agency. Thus, applied some ranges of RDX have been restricted as a result of soil and groundwater pollution (U.S. Environmental Protection Agency, 2017). Bacterial degradation for RDX which consider a xenobiotic compound has been detected. RDX biodegradation mechanism may be understated in the light of comprising and fusing unusual cytochrome P450 system of flavodoxin domain to the N terminus of a cytochrome P450 (CYP177A1, XplA). XplB system played a key role as RDX biodegradation through flavodoxin reduction which dedicated to RDX degradation in Rhodococcus rhodochrous strain 11Y and catalyzes the reductive denotation of RDX, in final, under aerobic and anaerobic condition, RDX ring was cleavage [3]. 16S rRNA gene-based specific primers consider powerful genetic biomarkers tools that used as indicators for potential contaminant biodegradation. Regarding to previous findings, applied biomarkers targeting conserved regions of 16S rRNA gene to identify microbial organisms on taxonomic and phylogenetic levels are effective when biodegradation is dependent on a specific microbial strain and has been widely accepted as a sensitive and reliable technique to identify intestinal micro biota [4-6]. This investigation was carried out to achieve two main goals. Firstly, identify bacterial genera with capability of RDX components biodegradation. Secondly, evaluate biodegradation activity for RDX components.
Material and Methods
Locations and sampling of soil
Soil samples were collected from four explosives contaminated sites at Riyadh province, Saudi Arabia (22.7554° N, 46.2092° E). Contaminated soil with explosives xenobiotic compounds had an average pH of 6.4, moisture content of 62.8%, conductivity of 290 μS per cm by contrary, few meters away from the TNT-contaminated location, non-contaminated soil had an average pH of 7.2, moisture content of 31%, conductivity of 378 μS per cm.
EPA-method 8510 (Nitroaromatics, Nitramines, and nitrate esters by High Performance Liquid Chromatography, HPLC) was applied for extraction of explosives from soil and Agilent 1100 system (Agilent, Santa Clara, CA) with Chromosphere C18 reverse phase column (5 μm, 250 × 4.6 mm) was applied. The method detection limit was approximately 2 µg/L for RDX. The detection limit was 5 µg/L for the RDX breakdown products hexahydro-3,5-dinitro-1-nitroso-1,3,5-triazine (MNX), hexahydro-5-nitro-1, 3-dinitroso-1,3,5-triazine (DNX), and hexahydro-1,3,5-trinitroso-1,3,5-triazine (TNX), TNT or related nitroaromatics were undetected in the non-contaminated soil (<0.01 ppm detection limit). Upper 5–10 cm of tested soil was applied via taken 50 grams. Pooling of soil samples was compounded from thee replicating to make one sample and kept at 4°C during transportation. Sieves (2 mm) were employed to homogenize the samples. Obtaining rhizosphere soil was achievement via shaking for 10 min at max. Speed in sterile P-buffer (per l: 2.36 g NaH2PO4; 1.80 g NaH2PO4, 85.0 g NaCl and 200 μl Tween 80; pH 6.8). The tubes were subjected to centrifugation (2,500 g, 20 min), resulting pellet was kept. All soil samples were stored at −80°C till DNA extrication.
Amplification, cloning and sequence of 16S rDNA
16S rRNA gene diversity analysis was employed to identify five bacterial which isolate from five explosive contaminated sites at Riyadh province, Saudi Arabia. Total genomic DNA was extracted and purified using E.Z.N.A.®Soil DNA Kit (D5625-01, OMEGA bio-tek, USA) according to manufacturer protocol. 16S rDNA fragments were amplified by PCR technique using 27F and 1492R primers (Table 1). Biobasics, Canada) according to DeLong [7]. PCR amplification was carried out in a themal cycler (peqSTAR 96X Universal Gradient, Germany) under conditions as follows: 5-min initial denaturation at 95°C, 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by 10 min at 72°C and incubation at 4°C [8]. Gel documentation system (Geldoc-it, UVP, England), was applied for data analysis using Total lab analysis software. The PCR amplicons were blunt-ended with Klenow polymerase and ligated to an Eco RV-digested pBluescript II plasmid (Stratagene) and transformed into Escherichia coli JM 109 electro competent cells according to the manufacturer's instruction. Transformed ligated cells were cultured on LB agar plates containing 100 mg/ml Ampicillin, 100 mg/ml IPTG, and 100 mg/ml X-Gal. Genomics DNA of transformed white colonies were sequenced by Applied Biosystems® 3730 DNA Analyzer. GenBank database was compared with 16S rRNA nucleotide sequences [9] through the BLASTn program [10] and identified with Ribosomal Database Project (RDP) [11] and Alignments sequences were analysed on NCBI website using BLAST to confirm their identity. The Genetic distances and Multi Alignments were computed by Pairwise Distance method using ClusteralW software analysis.
| RDX Degrading genes |
PRUN518R |
5′-ACTCCTACGGGAGGCAGCAG-3′; |
La Para [8] |
| PRBA338F |
5′-ATTACCGCGGCTGCTGG-3′ |
| Bacterial 16S rRNA |
xplB-KpnF1 |
5′-GGTACCATGGACATCATGAGT-3′ |
Chun Shiong Chong [12] |
| xplA-HindIIIF |
5′-AAGCTTATGACCGACGTAACTGTC-3′ |
Table 1: Primers for identification of 16S rRNA and RDX degrading genes.
Mining xpl RDX degrading genes
XplA and XplB (RDX degrading genes) encoding a novel flavodoxin fused cytochrome P450 were mining, amplifying and sequence through xplA-HindIIIF and xplB-KpnF1 specific primers (Table 1) from transformed bacterial cells, for xplA and xplB respectively. Thermo Scientific™ DreamTaq™ Hot Start Master Mix (Thermo Scientific, USA, K9011) was applied according to manufacturer protocol. PCR started with an initial denaturation step of 95°C for 5 minutes, followed by 30 cycles of denaturation step of 95°C (30 s), annealing step (varied temperature determined from primers melting temperature) and extension step of 72°C (1-3 minutes). A final extension cycle at 72°C for 10 minutes was performed [12].
Western analysis for xpl RDX degrading genes
Selected transformed colonies were cultured and total soluble proteins were purified via TriFast (Peqlab, VWR company, isolation of RNA, DNA and Protein simultaneously). Then, SDS Polyacrylamide gel electrophoresis (SDS-PAGE) with 12% T was performed to distinguish and fragment total soluble proteins. Nitrocellulose membrane (Schleicher and Shuell BioScience, Protran BA 83, cellulosenitrate (E), 0.2 μM, 200 × 200 mm, Dassel Germany) and filter paper (Whatman, 3 mm chromatography paper) were applied for performing western blotting technique. After protein transfer, membrane was incubated with anti-rabbit polyclonal antibodies were prepared by using purified XplA and XplB proteins, respectively, in rabbit [13] and XplB [14] for one hour and reaction was stopped by washing the membrane with TBS, 0.1% Tween 20, TBS and water according to Chong [12]. Data analysis was performed via ChemiDoc-It® Imager (UVP, England).
Results and Discussion
This investigation aimed to identify and evaluate biodegradation capability of RDX components for different bacterial isolates. To achieve these goals, 16 rRNA specific genes were applied for identified thirty nine isolates from different four explosive contaminated sites at Riyadh province, Saudi Arabia. As shown in Figure 1 all crude bacterial which were extracted and purified form four locations reflected clear 16 rRNA specific fragments of distinguishing ~ 950-base-pair. Then, the amplified 16S rDNA fragments were cloned and sequence. Thirty eight clones were successfully sequence. Sequence results indicated that, Proteobacterium, Burkholderia and Rhodococcus were the major dominant identified genera sequences among partial-sequence analyses of 38 clones from each four location libraries from four explosive contaminated sites at Riyadh province, Saudi Arabia. Table 2 showed identified bacterial to isolate from each location with the highest assigned homologous matches based on 16S rDNA sequences. Distinguish acceptance for 16S rDNA-based analysis has been founded for bacterial taxonomy and phylogenetic a number of 16S ribosomal data are being integrated into the Ribosomal Database Project [11].
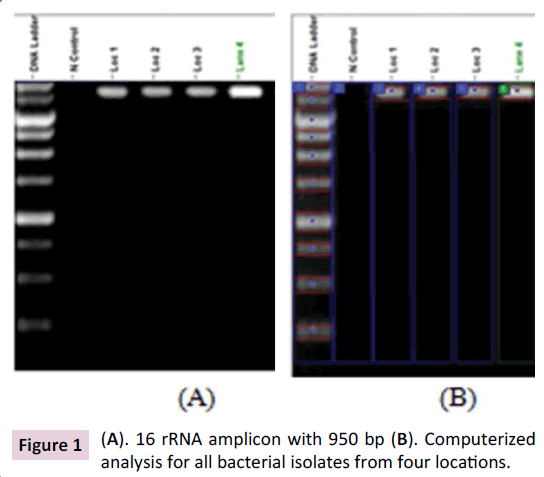
Figure 1: (A). 16 rRNA amplicon with 950 bp (B). Computerized analysis for all bacterial isolates from four locations.
| Location |
Strain |
Match of highest homology |
Accession
number |
| 1 |
1 |
Uncultured gamma proteobacterium isolate STB-666E |
EU330918.1 |
| 2 |
Uncultured gamma proteobacterium isolate STS-847E |
EU330898.1 |
| 3 |
Uncultured gamma proteobacterium isolate STB-666E |
EU330918.1 |
| 4 |
Uncultured gamma proteobacterium Isolate STS-847E |
EU330898.1 |
| 5 |
Uncultured gamma proteobacterium clone Ros119 |
AM237241.1 |
| 6 |
Uncultured gamma proteobacterium clone Ros102 |
AM237242.1 |
| 7 |
Uncultured gamma proteobacterium clone Ros30 |
AM237240.1 |
| 8 |
Burkholderia gladioli |
AB023647.1 |
| 9 |
Burkholderia gladioli |
AB023646.1 |
| 2 |
1 |
Burkholderia metallica strain R-16017 |
NR_042636.1 |
| 2 |
Uncultured beta proteobacterium clone Ros107 |
AM237275.1 |
| 3 |
Uncultured beta proteobacterium clone Ros12 |
AM237274.1 |
| 4 |
Uncultured beta proteobacterium clone Ros51 |
AM237273.1 |
| 5 |
Uncultured beta proteobacterium clone Ros59 |
AM237272.1 |
| 6 |
Uncultured beta proteobacterium clone Ros28 |
AM237268.1 |
| 7 |
Uncultured beta proteobacterium clone Ros78 |
AM237269.1 |
| 8 |
Uncultured beta proteobacterium clone Ros38 |
AM237264.1 |
| 9 |
Rhodococcus sp. KSM-B-3 |
AB032365.1 |
| 10 |
Rhodococcus sp. JFJ-Iso-Bact41 |
AJ867727.2 |
| 3 |
1 |
Burkholderia diffusa strain R-15930 |
NR_042633.1 |
| 2 |
Burkholderia alpina strain P0-03-17-38 |
JF_763852.1 |
| 3 |
Burkholderia alpina strain P0-04-17-38 |
NR_146371.1 |
| 4 |
Burkholderia thailandensis strain E264 |
NR_027587.1 |
| S |
Burkholderia semnalis strain R-2419 |
NR_042635.1 |
| 6 |
Burkholderia ubonensis strain GTC-P3-415 |
NR_040830.1 |
| 7 |
Enterobacter cloacae strain RJ30 |
KC990813.1 |
| 8 |
Enterobacter cloacae strain RJ20 |
KC990811.1 |
| 9 |
Enterobacter cloacae swain RJ15 |
AY017123.1 |
| 10 |
Enterobacter cloacae strain Vivi17 |
1 MF661959.1 |
| 4 |
1 |
Rhodococcus humicola strain UC33 |
NR_149271.1 |
| 2 |
Rhodococcus soli strain DSD51W |
NR _134799.2 |
| 3 |
Uncultured gamma proteobacterium |
AM237233.1 |
| 4 |
Enterobacter cloacae strain SEA01 |
KY625189.1 |
| 5 |
Burkholderia sp. MSMB122 |
KF378609.1 |
| 6 |
Uncultured beta proteobacterium clone Ro77 |
AM237261.1 |
| 7 |
Rhodococcus hoagii strain DSM 20307 |
NR_041910.1 |
| 8 |
Uncultured beta proteobacterium clone Ro35 |
AM237265.1 |
| 9 |
Rhodococcus sp. NCIB 9646 |
AY017136.1 |
Table 2: Bacterial isolates identification with assigned homologous matches.
Majority of bacterial genera for the first location was identified as proteobacterium, Burkholderia gladioli and Burkholderia metallica. Second location isolates was identified as different clones of β proteobacterium, Rhodococcus sp. KSM-B-3 and Rhodococcus sp. JFJ-Iso-Bact41. Different species of Burkholderia and Enterobacter were dominant for third location isolates. Fourth isolates location were identified generally as Rhodococcus, Burkholderia, β proteobacterium. Genetic similarity between thirty nine isolates and within location isolates were estimated and phyllogenetic tree was constructed for all studied locations as shown in Figure 2. More light was added to our findings through using 16S rRNA gene sequence similarity in identified bacterial genera was added [15]. They identified genus Comamonas based on 16S rRNA gene sequence search in Ribosomal Database Project, small subunit rRNA and large subunit rRNA databases together with the phyllogenetic tree analysis. Also, our finding was in agreements on Chong et al. [11]. They constructed Phyllogenetic analysis of the 16S rRNA gene sequences to compare the four RDX degrading Rhodococcus isolates. Also, for identified monooxygenase-associated genes in Pseudonocardia dioxanivorans CB1190 used as biomarkers to biodegrade 1,4-dioxane in pure cultures and environmental samples [6]. Likewise of our results of applied bacteria-specific primers to amplify the 16S rRNA genes to construct phylogenetic analysis for bacterial biodegradation of explosives in groundwater [16] Clostridium and Pseudomonas known explosive-degrading multiple sequences strains were identified and grouped. With agreement on dominance of α- and β-proteobacteria which isolated and identified from RDX-contaminated saturated soils samples [17] identified the same bacterial phyla using terminal restriction fragment length polymorphism (tRFLP) analysis of 16S rRNA genes.
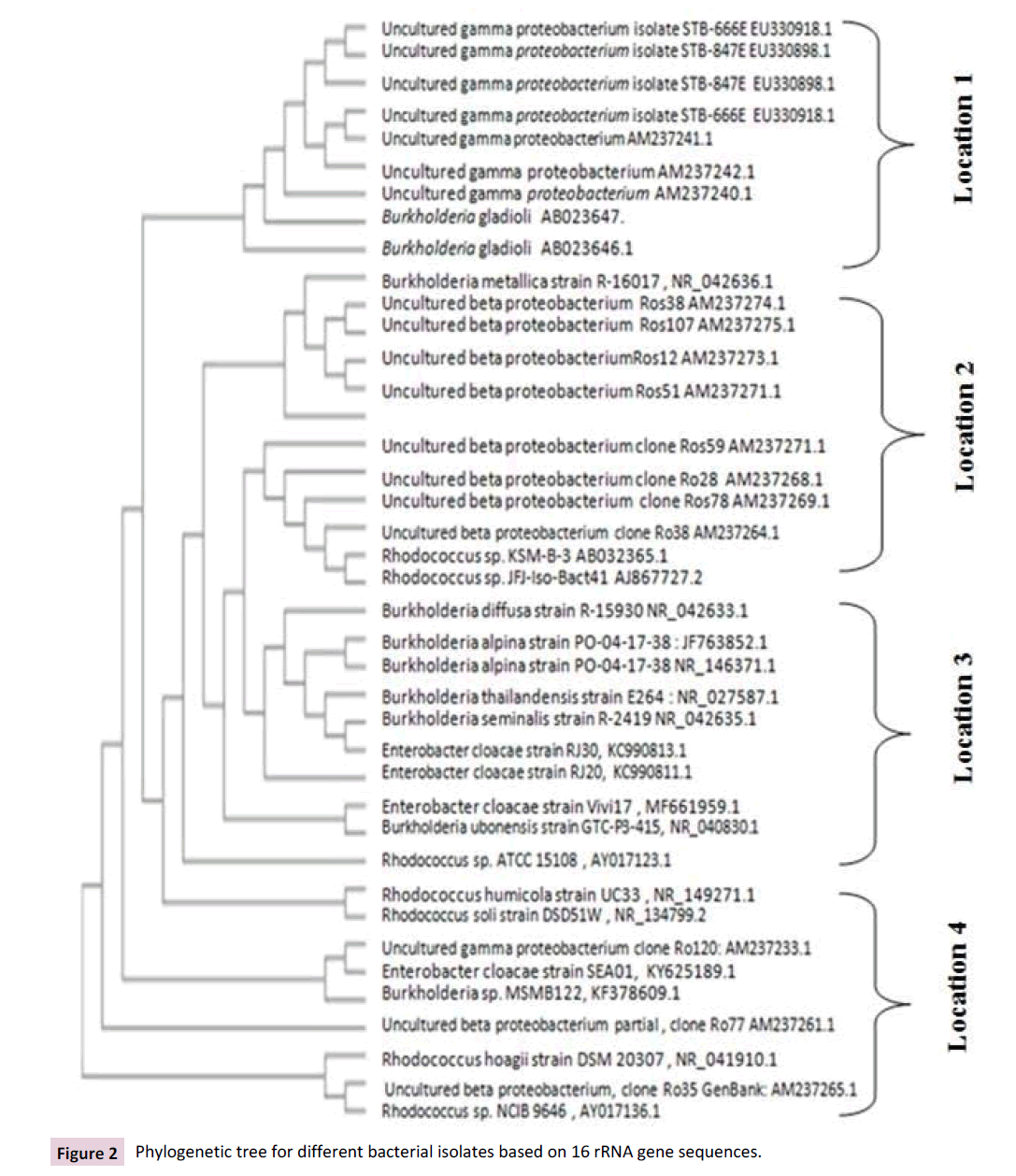
Figure 2: Phylogenetic tree for different bacterial isolates based on 16 rRNA gene sequences.
XplA and XplB (RDX degrading genes) were applied as molecular marker for evaluating RDX components biodegradation capability of previous identified isolates. As shown in Figure 3, specific differentiation for XplA and XplB proteins level were detected for thirty eight identified bacterial genera which isolated from four different locations. XplA and XplB biodegradation genes were only expressed for all first location isolates. Second location expressed XplA and XplB biodegradation genes for all isolates except identified bacterial isolates as Burkholderia metallica strain R-16017 (NR_042636.1) isolate. Interestingly, identified bacterial isolates as uncultured beta proteobacterium clone Ros107 (AM237275.1) was failed to express XplB biodegradation gene. Third isolate location reflect clear to distinguish expression profile for XplA and XplB genes, only bacterial isolates which identified as Burkholderia seminalis strain R-2419 (NR_042635.1) and Enterobacter cloacae strain Vivi17 (MF661959.1) suppressed XplA and XplB genes. Furthermore, XplB biodegradation gene was suppressed for bacterial isolating which identified as Burkholderia ubonensis strain GTC-P3-415 (NR_040830.1) and Enterobacter cloacae strain RJ30 (KC990813.1). Identify bacterial which isolates from fourth location reflected the lowest existence of XplA and XplB biodegradation genes comparing with other locations were Rhodococcus humicola strain UC33 (NR_149271.1) and Rhodococcus soli strain DSD51W (NR_134799.2) suppressed XplA and XplB genes. XplA gene was suppressed for uncultured gamma proteobacterium (AM237233.1) and Rhodococcus sp. NCIB 9646 (AY017136.1) and Burkholderia sp. MSMB122 (KF378609.1) suppressed XplB gene.
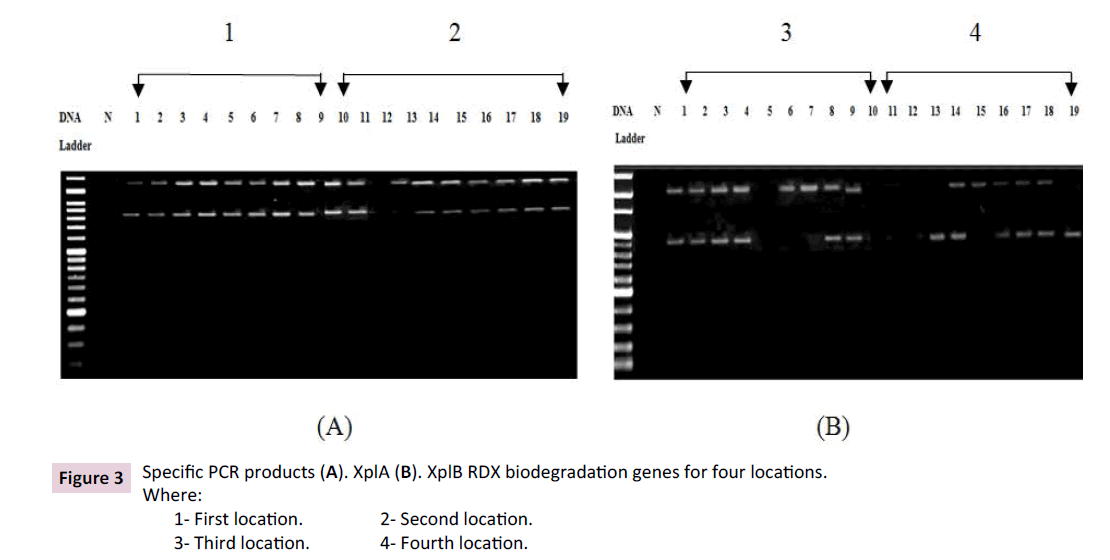
Figure 3: Specific PCR products (A). XplA (B). XplB RDX biodegradation genes for four locations.
Where:
1- First location. 2- Second location.
3- Third location. 4- Fourth location.
As shown in Figure 4, XplA and XplB proteins level were compared between bacterial isolates from varied locations which performed via western blotting technique indicated that, bacterial isolates from first location reflected the highest XplA and XplB proteins level and second; third and fourth bacterial isolates were arranged distinctly for XplA and XplB proteins level. Among first location isolates which were superior levels comparing with other locations, Burkholderia gladioli isolates (AB023647.1 and AB023646.1) showed the highest proteins level. On the other hand, Rhodococcus sp. KSM-B-3 (AB032365.1) and Rhodococcus sp. JFJ-Iso-Bact41(AJ867727.2) expressed the highest XplA and XplB proteins level comparing with rest isolate from second location. Third isolates location reflected highly distinguishable proteins level for Burkholderia diffusa strain R-15930 (NR_042633.1), Burkholderia alpina strain PO-03-17-38 (JF_763852.1) and Burkholderia alpina strain PO-04-17-38 (NR_146371.1). All bacterial isolates from fourth location showed the lowest proteins level for XplA and XplB genes. By contrary of our findings, xplA, hydA, onr, xenA, and xenB genes which associated with RDX degradation were disappeared in all of the groundwater RDX contaminant samples [16]. Our previous findings for quantifying XplA protein level through western blotting were supported by presented results of Chong [12]. Who detected only a single band with 47 kDa with XplA antibody cross-reactivity? One of important explanation for XplA and XplB expression level variation which detecting in our findings were cleared in the light of Chong [3] through applying Western blot analysis of crude extracts probed with XplA and XplB antibodies and indicated that that XplA and XplB protein levels were distinguishably decrease in. Also, open reading frame of xplB was not disrupted which cleared possibility for affection of mutations transcript stability or translational efficiency.
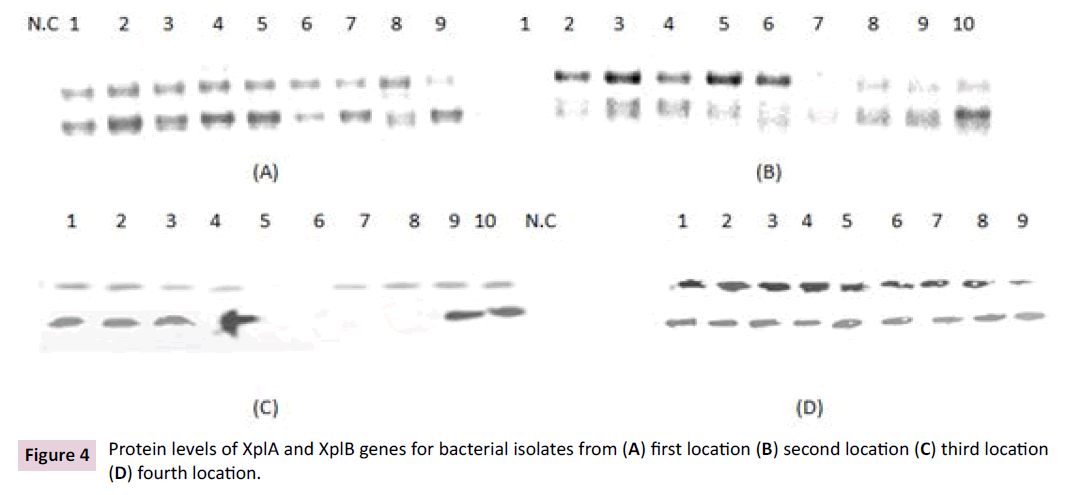
Figure 4: Protein levels of XplA and XplB genes for bacterial isolates from (A) first location (B) second location (C) third location (D) fourth location.
Conclusion
Our findings for identification and mining for biodegradation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) summarized that different bacterial genera with certain biodegradable activity were identified via 16S rRNA molecular marker technique. Furthermore, specific biodegradation genes, XplA and XplB showed different expression levels consistent with locations of bacterial isolation. Further studies were requirement to understand biodegradation molecular metabolic interaction for isolating from different locations.
Acknowledgement
This work was funded by the Italian Flagship Project NanoMax (S.P.)
References
- Rylott EL, Jackson RG, Sabbadin F, Seth-Smith HM, Edwards J, et al. (2011) The explosive-degrading cytochrome P450 XplA: biochemistry, structural features and prospects for bioremediation. Biochim Biophys Acta 1814: 230-236.
- Seth-Smith HMB, Edwards J, Rosser SJ, Rathbone DA, Bruce NC (2008) The explosive-degrading cytochrome P450 system is highly conserved among strains of Rhodococcus spp. Appl Environ Microbiol 74: 4550–4552.
- Chong CS, Sabir DK, Lorenz A, Bontemps C, Andeer P, et al. (2014). Analysis of the xplAB-containing gene cluster involved in the bacterial degradation of the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine. Appl Environ Microbiol 80: 6601-6610.
- Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R (2004) Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol 70: 7220-8
- Layton BA, Walters SP, Lam LH, Boehm AB (2010) Enterococcus species distribution among human and animal hosts using multiplex PCR. J Appl Microbiol 109: 539–547.
- Gedalanga P, Pornwongthong P, Mora R, Chiang SD, Baldwin B, et al. (2014) Identification of biomarker genes to predict biodegradation of 1,4-dioxane. Appl Environ Microbiol 80: 3209-3218.
- DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689.
- LaPara TM, Nakatsu CH, Pantea L, Alleman JE (2000) Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl Environ Microbiol 66: 3951–3959.
- Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BFF, et al. (1999) GenBank. Nucleic Acids Res 27: 12–17.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
- Cole JR, Wang Q, Fish JA, Chai A, McGarrell DM, et al. (2014) Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res 42: D633-D642.
- Chong CS (2011) Biodegradation of RDX in Rhodococcus spp. University of York Department of Biology, USA.
- Rylott EL, Jackson RG, Edwards J, Womack GL, Sethâ€ÂÂÂÂSmith HM, et al. (2006) An explosiveâ€ÂÂÂÂdegrading cytochrome P450 activity and its targeted application for the phytoremediation of RDX. Nat Biotechnol 24: 216–219.
- Jackson RG, Rylott EL, Fournier D, Hawari J, Bruce NC (2007) Exploring the biochemical properties and remediation applications of the unusual explosiveâ€ÂÂÂÂdegrading P450 system XplA/B. Proc. Natl Acad Sci USA 104: 16822–16827.
- Yadav V, Prakash S, Srivastava S, Verma PC, Gupta V, et al. (2009) Identification of Comamonas species using 16S rRNA gene sequence. Bioinformation 3: 381–383.
- Fuller ME, McClay K, Higham MP, Hatzinger PB, Steffan RJ (2010) Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) Bioremediation in Groundwater: Are Known RDX-Degrading Bacteria the Dominant Players? Bioremediation J 14: 121–134.
- Ringelberg DB, Reynolds CM, Walsh ME, Jenkins TF (2003) RDX loss in a surface soil under saturated and well drained conditions. J Environ Qual 32: 1244-1299.





