Keywords
Insecticide; Bombus terrestris; Neonicotinoid; Arylalkylamine N-acetyltransferase
Introduction
Ever-increasing insecticide resistance illustrates the uphill battle faced by the food and agricultural industries. Newly developed insecticides are quickly becoming classified as “non-renewable resources”, with a measure of effectiveness based on how long they will be commercially viable as a primary concern [1]. Adding to this concern is the link between current insecticide usage and declining bee populations [2]. Thus, the identification and exploitation of novel targets for insecticide design is critical to debase the detrimental effects of insecticide resistance and environmental damage. One such avenue, which holds potential, are the arylalkylamine N-acetyltransferases. Arylalkylamine N-acetyltransferases (AANAT), often called dopamine- or serotonin N-acetyltransferases, are members of the Gcn5-related N-acetyltransferase (GNAT) superfamily of enzymes. These enzymes catalyze the acetyl-CoA-dependent acetylation of an amine or arylalkylamine (Figure 1) [2].
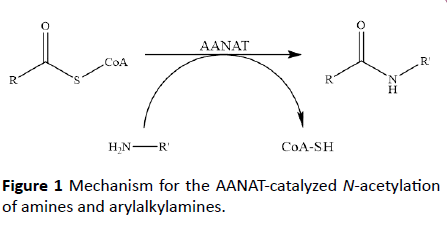
Figure 1: Mechanism for the AANAT-catalyzed N-acetylation of amines and arylalkylamines.
Literature Review
Since the initial characterization of Drosophila melanogaster AANAT by Maranda et al. [3], a large number of insect genes encoding AANAT and AANAT-like proteins have been described, highlighting the biological importance of these enzymes [4-10]. In vertebrates, AANAT catalyzes the rate-limiting step in the biosynthesis of melatonin [8] and has a key role in regulation of the sleep-wake cycle [9]. AANATs found in insects (iAANAT) are similarly involved in important biological processes, including insect-specific roles. It is a testament to the versatility of this group of enzymes that each insect family has developed family-unique AANATs to facilitate their own needs. iAANATs are vital to cuticle morphology by catalyzing the acetylation of dopamine to N-acetyldopamine, a precursor to the sclerotization, and the hardening of the exoskeleton [6,10]. In addition to dopamine, a variety of biogenic amine neurotransmitters also serve as acetyl group acceptors, meaning that the iAANATs are involved in neurotransmitter inactivation and, thus, may contribute to the regulation of neural signaling networks [11].
An iAANAT-targeted insecticide would not only disrupt neural signaling, but would also inhibit cuticle development, decrease the structural stability, result in a non-conforming appearance (harming the insects’ ability to mate), and diminish the cuticle-mediated protection against injury and infection. The aforementioned AANAT-regulated production of melatonin (by catalyzing the acetylation of serotonin) manages the life span of Drosophila melanogaster, illustrating the toxicity potential of these biogenic amines if their inactivation is interrupted. Insects are particularly susceptible to iAANAT inhibition because insects lack the enzyme monoamine oxidase, an enzyme which functions in mammals to inactivate arylalkylamines [12]. It has long been an endeavor of ours to examine the variety of functions of the iAANATs, and to ascertain the fatty acid amides they produce. Thus far, we have successfully identified, cloned and characterized iAANATs from Drosophila melanogaster and Bombyx mori. Other groups have characterized iAANATs from other insects, including Aedes aegypti, Tribolium castaneum and Antheraea pernyi [11,13-18]. A picture of diversity and specificity for iAANATs emerges from the kinetic analyses showing a broad range of amine and acyl-CoA thioester substrates for the iAANATs, knockdowns demonstrating the detrimental effects of targeting iAANATs, and sequence comparisons between the iAANATs revealing insect-specific targeting potential. These data point towards the iAANATs as excellent targets for the design of novel insecticides.
Aside from the biogenic amines which have shown cross-species functionality, acyl-CoA chain length specificity differs between specific iAANATs, ranging from acetyl-CoA to oleoyl-CoA. Based on the specificity of the acyl-CoA thioester substrates, some iAANATs function in the formation of both short- and long-chain N-acylamides, whereas others function only to catalyze the formation of short-chain or long-chain N-acylamides. For example, iAANATs found in D. melanogaster show wildly differing results, especially with regards to acyl-chain length for their respective acyl-CoA substrates. Acetyl-CoA is a substrate for D. melanogaster Dm-AANATA, Km = 39 ± 12 μM, but apparently had no appreciable affinity for the long-chain acyl-CoA, arachidonoyl-CoA [13]. Yet in the same organism, Dm-AANATL2 accepted both acetyl-CoA and arachidonoyl-CoA as substrates, with Km values of 6.1 ± 0.3 μM and 1.9 ± 0.25 μM, respectively, illustrating the differences in between the iAANATs found in the same organism [18]. This trend is similarly seen in comparing two iAANATs expressed by B. mori. Acyl-chain length had negligible effect on the Km values for the acyl-CoA substrates for Bm-iAANAT while Bm-iAANATL3 preferred acyl-CoA substrates with an acyl chain length of 2-10 carbon atoms, with longer chain acyl-CoA thioesters not acceptable as substrates, at all (Table 1).
| Acyl-CoA |
(Km) (μM) |
| Dm-AANATA |
Dm-AANATL2 |
Bm-iAANATL3 |
Bm-iAANAT |
| Acetyl-CoA |
39 ± 12 |
6.1 ± 0.27 |
90 ± 3.6 |
0.31 |
| Butyryl-CoA |
36 ± 2 |
1.8 ± 0.17 |
14 ± 0.75 |
N/A |
| Hexanoyl-CoA |
23 ± 3 |
N/A |
13 ± 0.54 |
N/A |
| Octanoyl-CoA |
18 ± 3 |
N/A |
10 ± 1.0 |
N/A |
| Decanoyl-CoA |
220 ± 60 |
N/A |
12 ± 5.5 |
N/A |
| Palmitoyl-CoA |
N/A |
9.9 ± 1.6 |
N/A |
1.1 ± 0.3 |
| Oleoyl-CoA |
N/A |
3.6 ± 0.58 |
N/A |
1.7 ± 0.51 |
| Arachidonyl-CoA |
N/A |
1.9 ± 0.25 |
N/A |
1.2 ± 0.67 |
Table 1: Km values for the iAANATs found in D. melanogaster and B. mori with respect to different acyl chain lengths.
A review of the data for the amine substrates for the iAANATs again point to specific metabolic functions for the specific members of the iAANAT family. Differences of several orders of magnitude are found for amine Km values when comparing iAANATs (Table 2). For example, the Km for dopamine with Dm-AANATA is 25 ± 2 μM and that for Bm-iAANATL3 is 330 ± 23 μM. Differences like this are congruent for numerous amines, signifying a divergence in binding specificity in the active site between iAANATs.
| Amines |
(Km) (μM) |
| Dm-AANATA |
Dm-AANATL7 |
Dm-AgmNAT |
Bm-iAANATL3 |
| Tyramine |
12 ± 1 |
42 ± 2 |
N/A |
63 ± 6.7 |
| Octopamine |
10 ± 2 |
120 ± 10 |
N/A |
18 ± 0.77 |
| Dopamine |
25 ± 2 |
170 ± 20 |
N/A |
330 ± 23 |
| Tryptamine |
33 ± 3 |
26 ± 2 |
N/A |
97 ± 8.8 |
| Norepinephrine |
32 ± 6 |
230 ± 50 |
N/A |
140 ± 8.6 |
| Phenethylamine |
56 ± 13 |
320 ± 40 |
N/A |
N/A |
| Serotonin |
110 ± 8 |
160 ± 20 |
N/A |
1100 ± 63 |
| 4-methoxyphenethylamine |
780 ± 60 |
190 ± 20 |
N/A |
N/A |
| 4-phenylbutylamine |
270 ± 20 |
610 ± 30 |
N/A |
N/A |
| 3,4-dimethoxyphenethylamine |
3200 ± 300 |
320 ± 30 |
N/A |
N/A |
| Histamine |
N/A |
520 ± 50 |
N/A |
1700 ± 98 |
| Putrescine |
N/A |
81 ± 11 |
51 ± 3 |
N/A |
| Agmatine |
N/A |
790 ± 100 |
0.3 ± 0.02 |
N/A |
| Spermine |
N/A |
N/A |
18 ± 3 |
N/A |
| N8-acetylspermine |
N/A |
N/A |
9.1 ± 1 |
N/A |
| Spermidine |
N/A |
N/A |
17 ± 0.5 |
N/A |
| Cadaverine |
N/A |
N/A |
32 ± 6 |
N/A |
Table 2: Binding constants for iAANATs found in D. melanogaster and B. mori with respect to different amine acceptors.
Results and Discussion
Kinetic analyses provide a limited evaluation of the active site differences between the iAANATs. The Km values are not Kdissociation values and a comparison (or ratio) of Km values may only provide an estimate ratio of Kdissociation values for an acyl-CoA or amine substrate. However, the highlighted differences in the Km values imply a large enough distinction in substrate affinity to suggest that insecticides can be developed that will only target iAANATs in insect pests. Another aspect in considering the iAANATs as insecticide targets is to evaluate expression knockdown data. If the expression knockdown of a potential insecticide target is not deleterious to the insect, an inhibitor targeting it will probably not be a useful insecticide. One such knockdown was performed by Long et al. on Bombyx mori iAANAT2 [18]. The inhibition of Bm-iAANAT2 expression led to an increase in melanin deposition in larvae and adults, resulting from increase cellular concentrations of dopamine. As mentioned, any deviation in the appearance of the insect would have a naturally detrimental effect on its chances of mating [19]. Another iAANAT knockdown was performed in T. castaneum, the red flour beetle, an insect regarded as a serious pest to the food industry and hence a perfect model for insecticide target discovery. Noh et al. [6] demonstrated that along with darkening of the cuticle, the knockdown resulted in a separated, misshapen elytron (the wing casing of the beetle), and misfolding of the hindwings of the beetle. The evidence demonstrates that targeting iAANAT compromised the structural integrity of the T. castaneum exoskeleton. This naturally leaves the insect vulnerable to a host of environmental threats, including disease, predation, and the aforementioned inability to mate.
While the experimental data point toward the iAANATs as excellent targets for insecticide development, insecticides have come under scrutiny for their toxicity against the honey bee, Apis melifera, and the bumblebee, Bombus terrestris. Neonicotinoid insecticides are particular offenders, claimed to be involved in colony collapse disorder (CCD) [20]. It is here the AANATs stand out as insecticide targets. An examination of the sequences for numerous iAANATs reveals several unique iAANAT families. The “traditional” iAANAT gene is easily identifiable through a CoA binding pocket motif (synonymous with the NAT superfamily) and an insect specific motif, FxDEPLN. This motif contains functionally and structurally important residues. Due to the link of the enzyme to dopamine acetylation, this “traditional” iAANAT is often referred to as dopamine N-acetyltransferase (DAT). DAT is conserved among all insects. However, genome mining suggests that most insects contain several other types of iAANATs, also with characteristic motifs. These other families of iAANAT generally exhibit very low sequence homology outside of their phylogenic grouping. Examples of this can be found in the mosquito family (namely Aedes and Culex) with greater than 80% homology among other family members. Compared against insect genomes outside of this however, there is generally less than 30% homology. To add to this, A. melifera and B. terrestris both contain an iAANAT specific for Apidae (bees). Coleoptera (beetles) contain several more. This is further illustrated in Figure 2.
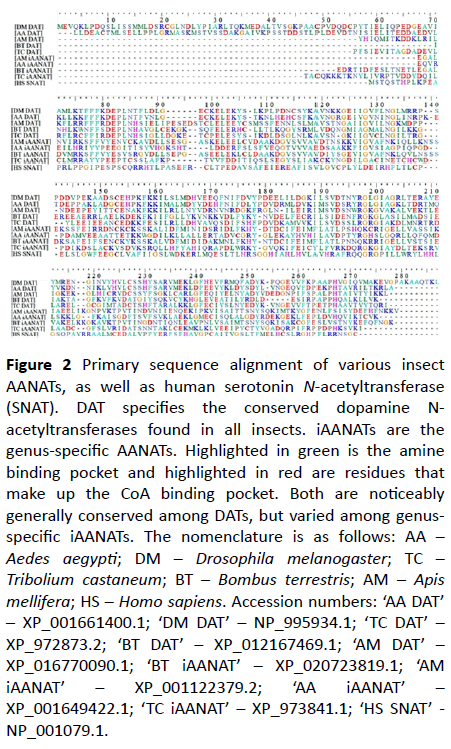
Figure 2: Primary sequence alignment of various insect AANATs, as well as human serotonin N-acetyltransferase (SNAT). DAT specifies the conserved dopamine N-acetyltransferases found in all insects. iAANATs are the genus-specific AANATs. Highlighted in green is the amine binding pocket and highlighted in red are residues that make up the CoA binding pocket. Both are noticeably generally conserved among DATs, but varied among genus-specific iAANATs. The nomenclature is as follows: AA – Aedes aegypti; DM – Drosophila melanogaster; TC – Tribolium castaneum; BT – Bombus terrestris; AM – Apis mellifera; HS – Homo sapiens. Accession numbers: ‘AA DAT’ – XP_001661400.1; ‘DM DAT’ – NP_995934.1; ‘TC DAT’ – XP_972873.2; ‘BT DAT’ – XP_012167469.1; ‘AM DAT’ – XP_016770090.1; ‘BT iAANAT’ – XP_020723819.1; ‘AM iAANAT’ – XP_001122379.2; ‘AA iAANAT’ – XP_001649422.1; ‘TC iAANAT’ – XP_973841.1; ‘HS SNAT’ - NP_001079.1.
Conclusion
Importantly, the regions responsible for binding of both the acyl-CoA and the amine substrate vary considerably between these iAANAT families, despite very high structural homology. In all iAANATs, the overall secondary and tertiary structure of the amine binding pocket is very similar. However, differences in specific residues are responsible for changes in binding affinities. In Diptera- (fly) specific iAANATs, for example, many hydrophobic residues are replaced with residues containing nucleophilic groups and hydrogen bond acceptors that are not found in other iAANATs [14]. This suggests that by exploiting these catalytically relevant, yet insect-specific residues, it is possible to design compounds that bind to a specific iAANAT family, such as Diptera iAANAT or Coleoptera iAANAT, which would not bind (or would bind with low affinity) to others, leaving non-targeted species unaffected.
These comparisons of iAANATs reveal the potential for high insecticide specificity, by solely targeting pests or disease vectors, and keeping agriculturally or ecologically important species unharmed. Additionally, iAANATs share little homology to AANATs expressed in vertebrates. This is in stark contrast to other types of insecticides, such as the aforementioned neonicotinoids, as well as organophosphates and organochlorines, which are highly non-specific, and often cause severe environmental side-effects [20]. Further work must be done to design and develop iAANAT-specific inhibitors to solidify these as legitimate targets. However, the evidence discussed herein indicates the iAANATs are excellent insecticide targets and the development of iAANAT-targeted inhibitors has the potential to solve many of the problems faced by current insecticide development.
Acknowledgements
This work has been supported, in part, by grants from the University of South Florida (a Creative Scholarship grant), the Shirley W. and William L. Griffin Charitable Foundation, and the National Institute of General Medical Science of the National Institutes of Health (R15-GM107864) to D.J.M.
Competing Interests
The authors declare no competing interests.
Authors’ Contributions
B. G. O., A. J. H., and D. J. M. contributed equally to the creation of this article.
References
- Barrès B, Micoud A, Corio-Costet MF, Debieu D, Fillinger S, et al. (2016) Trends and challenges in pesticide resistance detection. Trends Plant Sci. 2016 Oct 21: 834-53.
- Van der Zee R, Gray A, Pisa L, De Rijk T (2015) An Observational Study of Honey Bee Colony Winter Losses and Their Association with Varroa destructor, Neonicotinoids and Other Risk Factors. PLoS ONE 10: e0131611.
- Dyda F, Klein DC, Hickman AB (2009) GCN5-related N-acetyltransferases: A structural overview. Annu Rev Biophys Biomol Struct 29: 81-103.
- Ichihara N, Okada M, Takeda M (2001) Characterization and purification of polymorphic arylalkylamine N-acetyltransferase from the American cockroach, Periplaneta Americana. Insect Biochem Mol Biol 32: 15-22.
- Dai F, Qiao L, Tong X, Cao C, Chen P, et al. (2010) Mutations of an arylalkylamine-N-acetyltransferase, Bm-iAANAT, are responsible for silkworm melanism mutant. J Biol Chem 285: 19553–19560.
- Tsugehara T, Imai T, Takeda M (2013) Characterization of arylalkylamine N-acetyltransferase from silkmoth (Antheraea pernyi) and pesticidal drug design based on the baculovirus-expressed enzyme. Comp Biochem Physiol Part C Toxicol Pharmacol 157: 93-102.
- Dempsey DR, Jeffries KA, Bond JD, Carpenter AM, Rodriguez-Ospina S, et al. (2014) Mechanistic and structural analysis of Drosophila melanogaster arylalkylamine N-acetyltransferases. Biochem 53:7777-7793.
- Dempsey DR, Jeffries KA, Handa S, Carpenter AM, Rodriguez-Ospina S, et al. (2015) Mechanistic and structural analysis of a drosophila melanogaster enzyme, arylalkylamine n-acetyltransferase Like 7, an enzyme that catalyzes the formation of n-acetylarylalkylamides and N-acetylhistamine. Biochem 54: 2644-2658.
- Dempsey DR, Carpenter AM, Ospina SR, Merkler DJ (2015) Probing the chemical mechanism and critical regulatory amino acid residues of Drosophila melanogaster arylalkylamine N-acyltransferase like 2. Insect Biochem Mol Biol 66: 1-12.
- Noh MY, Koo B, Kramer KJ, Muthukrishnan S, Arakane Y (2016) Arylalkylamine N-acetyltransferase 1 gene (TcAANAT1) is required for cuticle morphology and pigmentation of the adult red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol 79: 119-129.
- Dempsey DR, Nichols DA, Battistini MR, Pemberton O, Ospina SR, et al. (2017) Structural and mechanistic analysis of Drosophila melanogaster Agmatine N-Acetyltransferase, an enzyme that catalyzes the formation of N-acetylagmatine. Sci Rep 7: 13432.
- Ganguly S, Coon SL, Klein DC (2002) Control of melatonin synthesis in the mammalian pineal gland: The critical role of serotonin acetylation. Cell Tissue Res 1: 127-137.
- Iuvone PM, Tosini G, Pozdeyev N, Haque R, Klein DC, et al. (2005) Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog Retin Eye Res 4: 433-456.
- Itoh MT, Hattori A, Nomura T, Sumi Y, Suzuki T (1995) Melatonin and arylalkylamine N-acetyltransferase activity in the silkworm, Bombyx mori. Mol Cell Endocrinol 115: 59-64.
- Sloley BD (2004) Metabolism of monoamines in invertebrates: The relative importance of monoamine oxidase in different phyla. NeuroToxicology 25: 175-183.
- Han Q, Robinson H, Ding H, Christensen BM, Li J (2012) Evolution of insect arylalkylamine N-acetyltransferases: Structural evidence from the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci 109: 11669-11674.
- Battistini MR (2015) Novel enzyme perspectives: Arylalkylamine N-acyltransferases from Bombyx mori and 1-Deoxy-D-Xylulose-5-Phosphate Synthase from Plasmodium falciparum and Plasmodium vivax [dissertation]. University of South Florida, USA.
- Long Y, Li J, Zhao T, Li G, Zhu Y (2015) A new Arylalkylamine N-Acetyltransferase in Silkworm (Bombyx mori) affects integument pigmentation. Appl Biochem Biotechnol 175: 3447-3457.
- Chung H, Carroll SB. Wax (2015) Sex and the origin of species: Dual roles of insect cuticular hydrocarbons in adaptation and mating. Bioessays 7: 822-830.
- Aktar MW, Sengupta D, Chowdhury A (2009) Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip Toxicol 2: 1-12.



