Naveeda Riaz1, Anum Shahbaz1 and Saima kalsoom2*
1Department of Bioinformatics and Biotechnology, International Islamic University, Islamabad, Pakistan
2(SA-CIRBS), Faculty of Basic and Applied Sciences, International Islamic university, Islamabad, Pakistan
Corresponding Author:
Saima kalsoom
(SA-CIRBS), Faculty of Basic and Applied Sciences
International Islamic university
Islamabad, Pakistan
E-mail: genious_chemist@yahoo.com
Received Date: March 06, 2018; Accepted Date: June 01, 2018; Published Date: June 09, 2018
Citation: Riaz N, Shahbaz A, Kalsoom S (2018) Ligand Based Pharmacophore Model Development for the Identification of Novel Anti-Psychotic Drugs. Appl Sci Res Rev 5:10. doi: 10.21767/2394-9988.100075
Keywords
Ligand scout; Bipolar disorder (BP); Antipsychotic; Pharmacophore
Introduction
Bipolar disorder is a chronic psychiatric illness characterized by depression and at least 1 manic or hypomanic episode during the lifetime of the illness [1]. In this disease illness vary from mild depression to severe form of mania, and effect both sexes equally., environmental and neurochemical factors are responsible for the prognosis of this disease. 3 major neurochemicals involve are serotonin, dopamine and noradrenaline. It occurs in specific part of brain and induce depression and mania as specific symptoms. Misspecification of who has a disease (low specificity) is generally more damaging to scientific inference than under detection (low sensitivity) [2]. 29 standard drugs were selected in this research having anti-depressant and anti-psychotic activities. Many drugs have been marketed for the treatment of bipolar disorder. Using pharmacophoric patterns comparative analysis of these drugs was checked. As a pharmacophore model should be able to discriminate between molecules with and without bioactivity, the set of molecules should include both active and inactive compounds. Different classes of compounds called antipsychotic, anticonvulsant, selective serotonin reuptake inhibitors (SSRIs), serotonin norepinephrine reuptake inhibitors (SNRIs), norepinephrine and dopamine reuptake inhibitors (NDRIs), noradrenergic and specific serotonergic antidepressants (NASSAs), Cyclics, and monoamine oxidase inhibitors (MAOIs) were studied and their structures were also obtained by using ChemDraw along with the LogP, molar refractivity (MR), critical volume (CV) and molecular weights (MW) values. Out of 29 compounds only 20 compounds were extracted for drug-likeness evaluation.
Pharmacophore models are useful for designing lead structure and identified also for Binding explaining site (Khan et al.). Ligand-based methods use only ligand information for predicting activity depending on its similarity/dissimilarity to previously known active ligands. It relies on knowledge of other molecules that bind to the biological target of interest, which may be used to derive a pharmacophore model that will define the minimum necessary structural characteristics a molecule must possess to bind to the target. It relies on knowledge of other molecules that bind to the biological target of interest, which may be used to derive a pharmacophore model that will define the minimum necessary structural characteristics a molecule must possess to bind to the target. A Pharmacophore may be defined as the essential geometric arrangement of atoms or functional groups necessary to produce a given biological response. IUPAC defines pharmacophore as: the ensemble of steric and electronic features that is necessary to ensure the optimal supramolecular interactions with a specific biological target structure and to trigger (or to block) its biological response. Recent pharmacophore models can be classified into two categories: receptor-based pharmacophores and ligand-based pharmacophores. For a receptor with a known three-dimensional structure, receptor-based pharmacophores have been studied which are based on the famous concept of a key for the lock [3]. Pharmacophore models are useful for designing lead structure and for explaining possible interactions of an identified binding site. Many investigations indicated that the presence of at least one Hydrophobic Unit, One Or two electron donor atoms, and/ or an NH group in as special spatial arrangement seems to be necessary in the structure of anticonvulsants [4]. In recent years, technological advances in virtual screening methodologies have allowed medicinal chemists to rapidly screen drug libraries for therapeutic activity against new biomolecular targets in a costeffective manner [5]. Adverse effects are the leading cause of concern in any medicated regimens [6]. In silico approaches can help us to design drugs with more efficacy and less side effects.
Methodologies
Drawing of structures
The structures of 29 compounds were drawn using Chem Draw Ultra 8.0 (Cambridge soft Corp (www.cambridgesoft.com), USA) and saved on MOL format. They are shown in the Table 1. Twenty compounds were taken out of 29 for further evaluation.
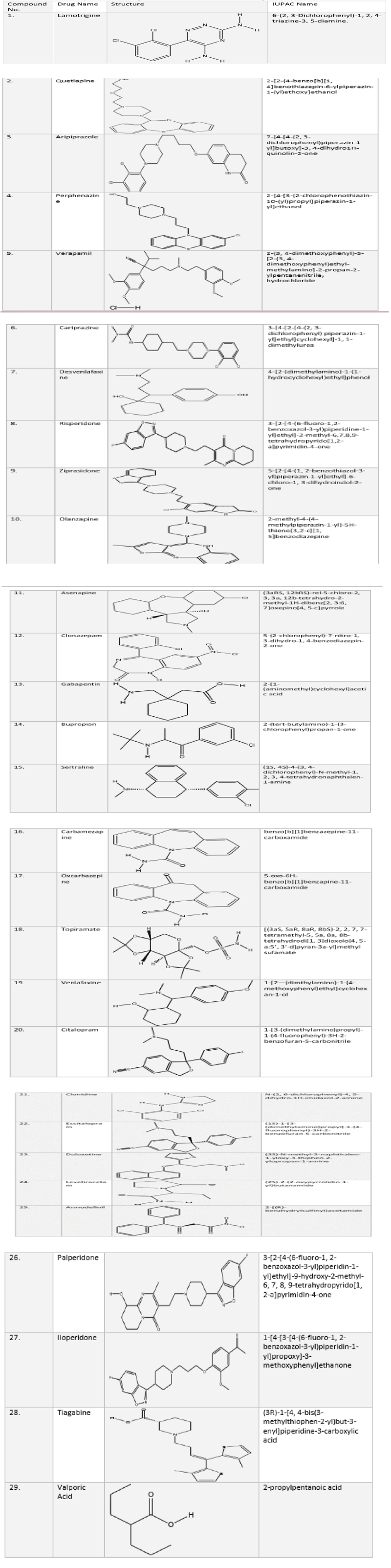
Table 1: List of compounds showing IUPAC names and structures.
Pharmacophore generations
Ligand scout is a software tool that allows to rapidly and transparently derive 3D pharmacophores from structural data of ligands in a fully automated and convenient way. The algorithms are scientifically published and based on several years of experience in pharmacophore creation [7,8].
There are some general steps that should be followed for this activity the working. Such as:
• Input a set of collected compound structures of drug-like molecules that are known to bind the receptor.
• These compounds were drawn on ChemDraw and saved in MOL file.
• All the parameters were set to default.
• Once the pharmacophore of all compounds was identified, the ligands are then superimposed.
• Superimposition allows the pharmacophore elements to overlap and a common template i-e the pharmacophore model is identified [9-13].
The pharmacophore model will then be tested using a validation set of known pharmacophore compounds available for bipolar disorder.
Visualization and distance calculation
VMD is designed for modeling, visualization, and analysis of biological systems such as proteins, nucleic acids, lipid bilayer assemblies, etc. VMD provides a wide variety of methods for rendering and coloring a molecule: simple points and lines, CPK spheres and cylinders, licorice bonds, backbone tubes and ribbons, cartoon drawings, and others [14-20].
Result and Discussion
Pharmacophore
Pharmacophore was defined by Peter Gund: “a set of structural features in a molecule that is recognized at a receptor site and is responsible for that molecule's biological activity”. This modern definition is remarkably loyal to the earliest definitions. Perceiving a pharmacophore is the first essential step towards understanding the interaction between a receptor and a ligand. (Tables 2 and 3) once a pharmacophore is established, a beneficial use of it is 3D database searching to retrieve novel compounds that would match the pharmacophore, without necessarily duplicating the topological features of known active compounds (hence remain independent of existing patents) (Figures 1-5).
| Compound No. |
No. of HBA |
No. of HBD |
No. of Ar |
No. of Hyd |
| 1 |
3 |
2 |
2 |
3 |
| 2 |
4 |
1 |
4 |
2 |
| 3 |
2 |
1 |
3 |
4 |
| 4 |
2 |
1 |
3 |
3 |
| 5 |
5 |
0 |
3 |
3 |
| 6 |
1 |
1 |
2 |
3 |
| 7 |
2 |
2 |
2 |
2 |
| 8 |
5 |
0 |
4 |
3 |
| 9 |
2 |
1 |
4 |
2 |
| 10 |
1 |
1 |
3 |
3 |
| 12 |
4 |
1 |
2 |
3 |
| 14 |
1 |
1 |
2 |
4 |
| 16 |
1 |
1 |
2 |
3 |
| 17 |
2 |
1 |
2 |
2 |
| 19 |
2 |
1 |
2 |
2 |
| 21 |
3 |
0 |
3 |
3 |
| 22 |
3 |
0 |
3 |
4 |
| 23 |
1 |
1 |
4 |
3 |
| 25 |
2 |
1 |
2 |
2 |
| 26 |
6 |
1 |
4 |
3 |
Table 2: 3D Pharmacophoric features of selected compounds showing number of hydrogen bond acceptor, hydrogen bond donor, aromatic rings and hydrophobic units against antipsychotic drugs.
| Compound No |
Ar1-Ar2 (nm) |
Ar1-HBA |
Ar2-HBA |
Ar1-Hyd (nm) |
Ar2-Hyd (nm) |
HBA-Hyd |
| |
|
(nm) |
(nm) |
|
|
(nm) |
| 1 |
6.95 |
5.92 |
3.9 |
3.9 |
4.76 |
6.35 |
| 2 |
6.73 |
5.77 |
3.28 |
3.87 |
4.97 |
6.09 |
| 3 |
6.59 |
5.48 |
3.56 |
3.42 |
4.51 |
6.26 |
| 4 |
6.67 |
5.11 |
3.58 |
3.92 |
4.39 |
6.05 |
| 5 |
6.7 |
5.13 |
3.3 |
3.25 |
4.29 |
6.07 |
| 6 |
6.52 |
5.58 |
3.8 |
3.91 |
4.4 |
6.59 |
| 7 |
6.32 |
5.98 |
3.64 |
3.87 |
4.83 |
6.44 |
| 8 |
6.66 |
5.88 |
3.7 |
3.4 |
4.8 |
6.3 |
| 9 |
6.84 |
5.9 |
3.46 |
3.86 |
4.27 |
6.52 |
| 10 |
6.74 |
5.47 |
3.76 |
3.23 |
4.54 |
6.75 |
| 12 |
6.41 |
5.9 |
3.58 |
3.41 |
4.64 |
6.36 |
| 14 |
6.23 |
5.5 |
3.6 |
3.84 |
4.74 |
6.5 |
| 16 |
6.92 |
5.75 |
3.81 |
3.89 |
4.95 |
6.42 |
| 17 |
6.3 |
5.9 |
3.6 |
3.27 |
4.38 |
6.29 |
| 19 |
6.2 |
5.94 |
3.78 |
3.39 |
4.47 |
6.59 |
| 21 |
6.45 |
5.86 |
3.3 |
3.25 |
4.28 |
6.08 |
| 22 |
6.55 |
5.1 |
3.86 |
3.84 |
4.57 |
6.74 |
| 23 |
6.82 |
5.72 |
3.71 |
3.33 |
4.96 |
6.35 |
| 25 |
6.24 |
5.95 |
3.42 |
3.91 |
4.36 |
6.58 |
| 26 |
6.48 |
5.25 |
3.56 |
3.44 |
4.87 |
6.66 |
Interactions of different compounds have been observed.
Table 3: Pharmacophoric distances of each twenty compounds.
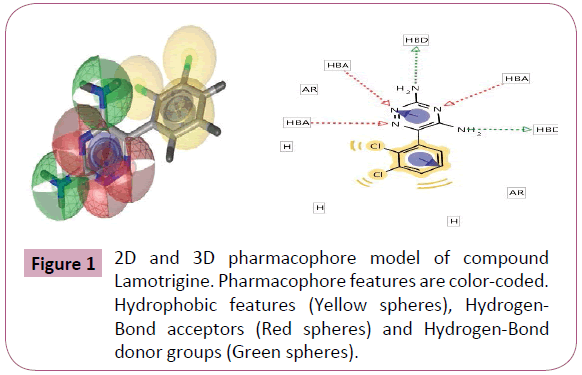
Figure 1: 2D and 3D pharmacophore model of compound Lamotrigine. Pharmacophore features are color-coded. Hydrophobic features (Yellow spheres), Hydrogen- Bond acceptors (Red spheres) and Hydrogen-Bond donor groups (Green spheres).
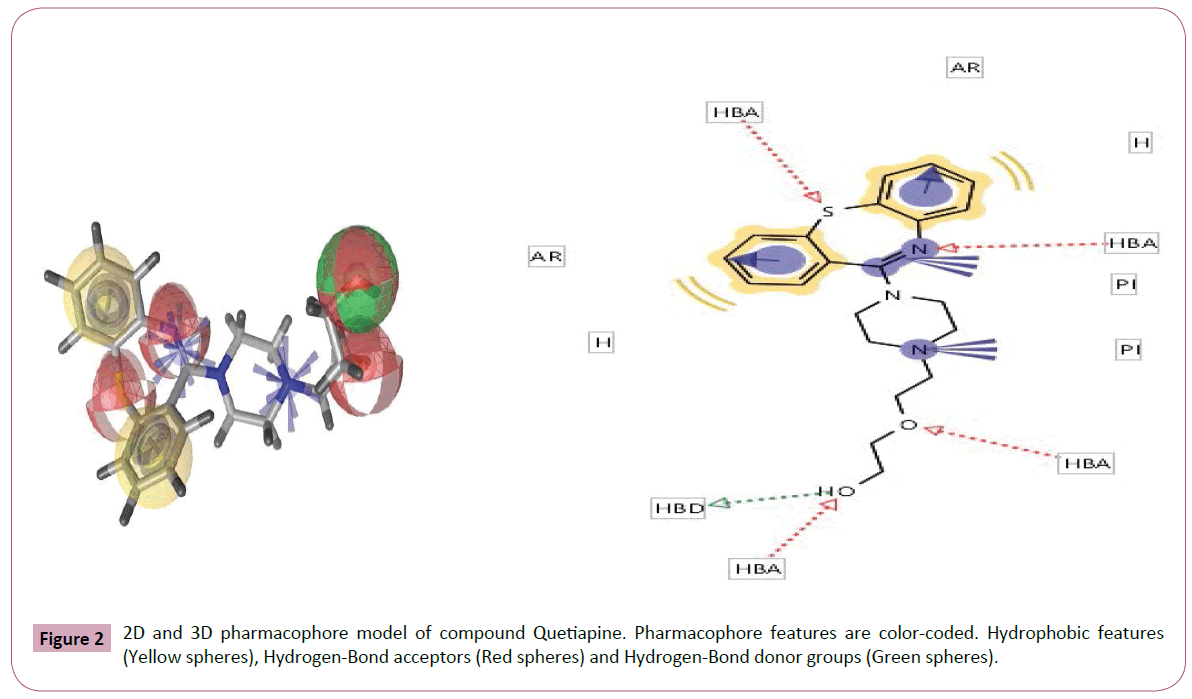
Figure 2: 2D and 3D pharmacophore model of compound Quetiapine. Pharmacophore features are color-coded. Hydrophobic features (Yellow spheres), Hydrogen-Bond acceptors (Red spheres) and Hydrogen-Bond donor groups (Green spheres).
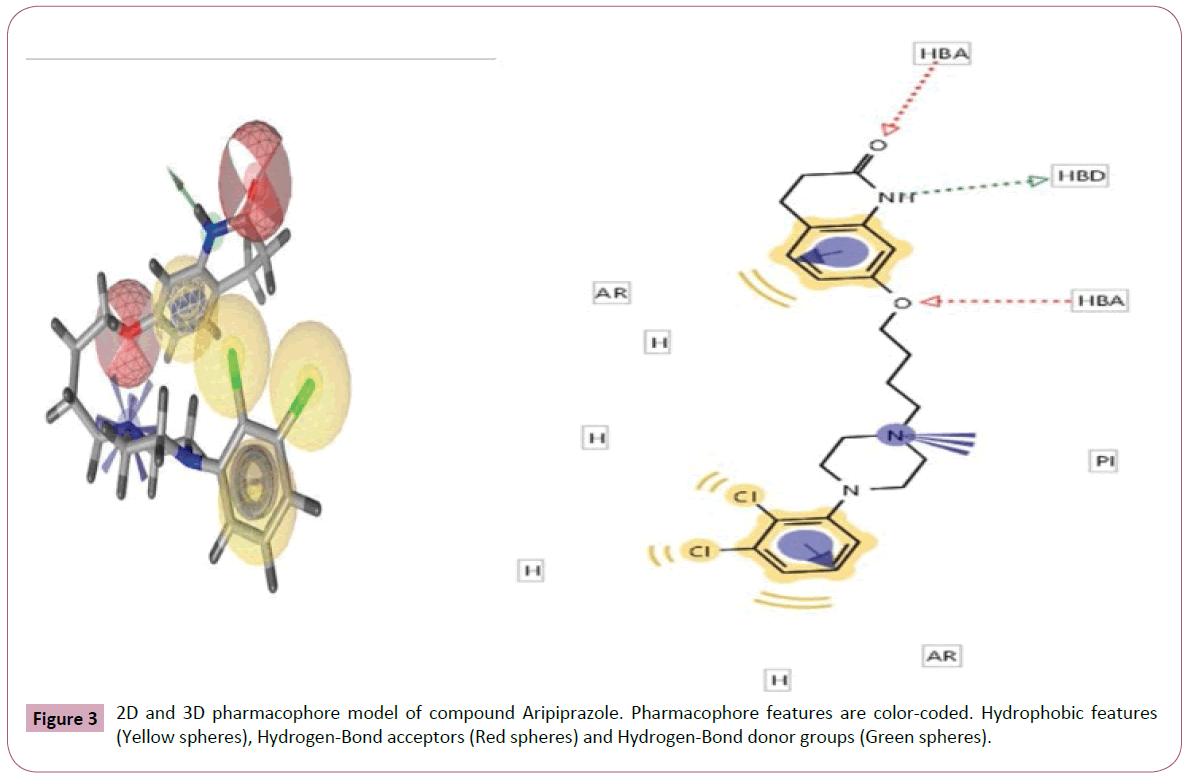
Figure 3: 2D and 3D pharmacophore model of compound Aripiprazole. Pharmacophore features are color-coded. Hydrophobic features (Yellow spheres), Hydrogen-Bond acceptors (Red spheres) and Hydrogen-Bond donor groups (Green spheres).
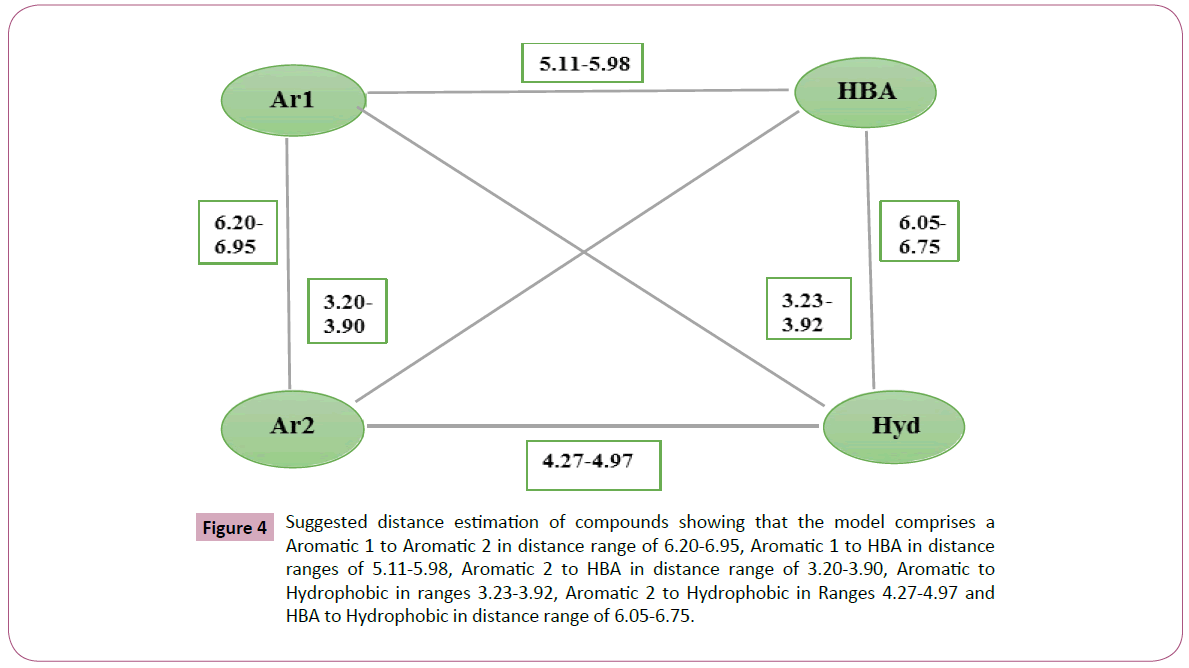
Figure 4: Suggested distance estimation of compounds showing that the model comprises a Aromatic 1 to Aromatic 2 in distance range of 6.20-6.95, Aromatic 1 to HBA in distance ranges of 5.11-5.98, Aromatic 2 to HBA in distance range of 3.20-3.90, Aromatic to Hydrophobic in ranges 3.23-3.92, Aromatic 2 to Hydrophobic in Ranges 4.27-4.97 and HBA to Hydrophobic in distance range of 6.05-6.75.
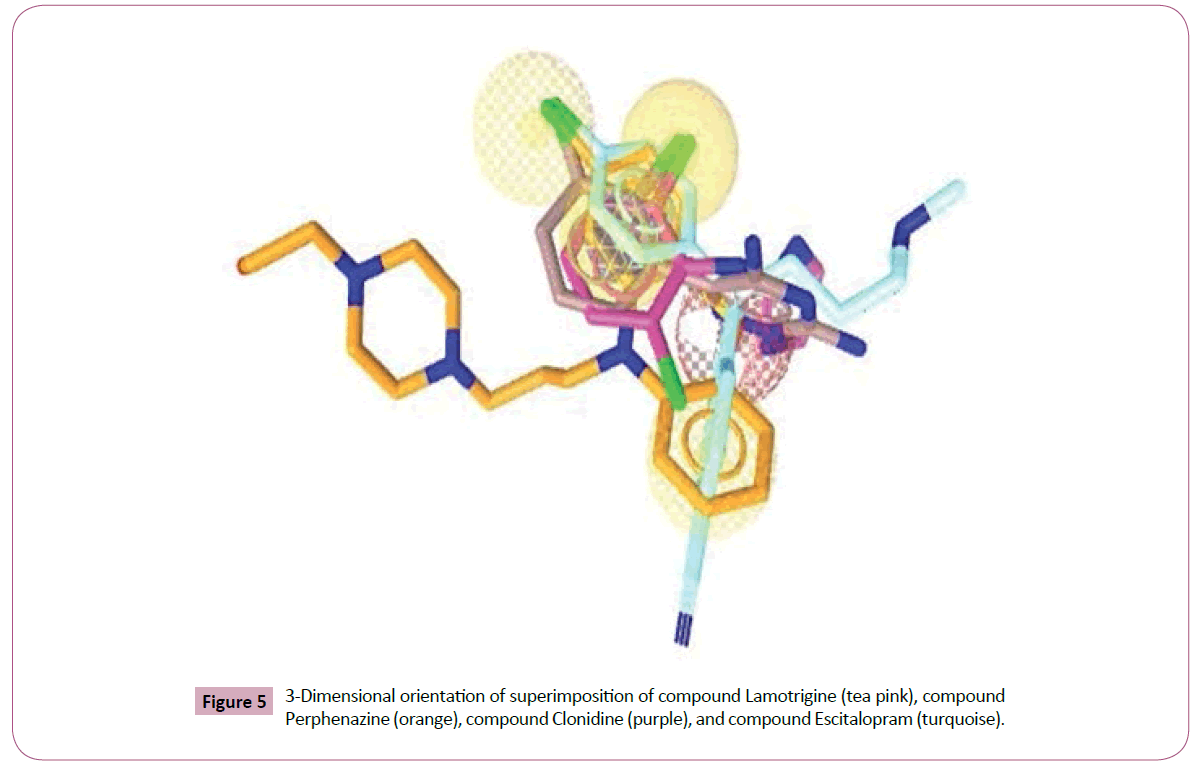
Figure 5: 3-Dimensional orientation of superimposition of compound Lamotrigine (tea pink), compound Perphenazine (orange), compound Clonidine (purple), and compound Escitalopram (turquoise).
Lipinski’s rule of five
The Lipinski’s rule of five contains certain criteria of features of compounds in specific amount. The Table 4 shows the general form of Lipinski’s rule of five.
| Properties |
Range |
| HBA |
= 6 |
| Aromatic |
= 4 |
| Hydrophobic |
= 4 |
| Log P |
<5 |
| Mol wt. |
<500 |
In this study 26 compounds were observed under the fulfillment of Lipinski’s rule of five.
Table 4: Lipinski’s Rule of five showing general form.
Conclusion
Pharmacophore model can be a multipurpose tool which aids in the discovery and development of new lead compounds. In this study, pharmacophore model for 20 new chemical compounds were validated. A ligand-based pharmacophore approach is used for the prediction of antipsychotic activity to get the compounds that are biologically active, and this eventually leads to an antipsychotic drug which is done using Ligand Scout. The general pharmacophore consisted of three main features namely, hydrophobic unit, hydrogen bonding acceptor and aromatic rings. The derived pharmacophore models are then filtered using Lipinski’s rule of five and orally bio-available compounds were obtained. These compounds will be use for the treatment of bipolar disorders.
References
- Hirschfeld RM, Calabrese JR, Weissman MM, Reed M, Davies MA, et al. (2003) Screening for bipolar disorder in the community. J Clin Psychiatry 64: 53-59.
- Franco EL, Franco ED, Rohan TE (2002) Evidence-based policy recommendations on cancer screening and prevention. ACRM 26: 350-361.
- Brooijmans N, Kuntz ID (2003) Molecular recognition and docking algorithms. Annu Rev Biophys Biomol Struct 32: 335-373.
- Camerman N (1980) Stereochemical similarities in chemically different antiepileptic drugs. Antiepileptic Drugs: Mechanisms of Action Raven Press, New York, pp: 223-231.
- Ma DL, Chan DS, Leung CH (2013) Drug repositioning by structure-based virtual screening. Chem Soc Review 42: 2130-2141.
- Wakaskar RR (2017) Challenges pertaining to adverse effects of drugs. Int J Drug Dev Res 9: 1-2.
- Wolber G, Seidel T, Bendix F, Langer T (2008) Molecule-pharmacophore super positioning and pattern matching in computational drug design. Drug Discov Today 13: 23-29.
- Wolber G, Langer T (2005) Ligand Scout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening ï¬ÂÂÂÂlters. J Chem Inf Model 45: 160-169.
- Belizario GO, Gigante AD, Rocca CCA, Lafer B (2017) Cognitive impairments and predominant polarity in bipolar disorder: a cross-sectional study. Int J Bipol Dis 5: 15.
- Wakaskar RR (2017) Challenges pertaining to adverse effects of drugs. Int J Drug Dev Res 9: 1.
- Guner OF (2002) History and evolution of pharmacophore in computer aided drug design. Curr Top Med Chem 2: 1321-1332.
- Hales ER, Kathleen TB, Hilty MD (1999) A review of bipolar disorder among adults. Psych Ser 50: 201-213.
- Hina NK , Saima K, Hamid R (2012) Ligand based pharmacophore model development for the identification of novel antiepileptic compound. Epi Res 98: 62-71.
- Kurogi Y, Guner OF (2001) Pharmacophore modelling and three-dimensional database searching for drug design sing catalyst. Current Med Chem 8: 1035-1055.
- Lundervold AJ, Heimann M, Manger T (2008) Department of biological and medical psychology. Br J Educ Psychol 78: 567-580.
- Mamdani F, Groisman IJ, Alda M, Turecki G (2004) Pharmacogenetics and bipolar disorder. Pharmacogenomics J, pp: 161-170.
- Manji KH, Quiroz AJ, Payne LJ, Singh J, Lopes PB, et al. (2003) The underlying neurobiology of bipolar disorder. World Psychia 3: 136-146.
- Morris GM, Lim-Wilby M (2008) Molecular docking. PubMed, pp: 82-26.
- Schneider MR, DelBello MP, McNamara RK, Strakowski SM, Adler CM (2012) Neuro progression in bipolar disorder. Bipolar Disord 14: 356-374.
- Markowitz, David J, Frank, Ellen, George, et al. (1996) Clinical trials-bipolar disorder. Psychopharmacol Bulletin 32: 613-621.