Short Communication - (2017) Volume 2, Issue 4
Chakraborty AK*
Department of Biotechnology and Biochemistry, Oriental Institute of Science and Technology (OIST), Vidyasagar University, West Bengal, India
*Corresponding Author:
Dr. Asit Kumar Chakraborty
Department of Biotechnology and Biochemistry
Oriental Institute of Science and Technology (OIST)
Vidyasagar University, West Bengal-721102, India.
Tel: (91) 9339609268
E-mail: chakraakc@gmail.com
Received Date: October 09, 2017; Accepted Date: October 24, 2017; Published Date: October 27, 2017
Citation: Chakraborty AK (2017) Mechanisms of AMR: Bacteria Won the Battle Against Antibiotics. Insights Biomed. Vol.2 No.4:18 DOI: 10.21767/2572-5610.100034
Antibiotic was discovered in 1926 by Alexander Flaming but only in 1943 after World War-II, penicillin was marketed for all followed by tetracycline and streptomycin. However, high dose uses of antibiotics cause rapid destruction of gut microbiota that help human growth and metabolism by providing vitamins and many other complex biomolecules. It appears that intestinal cells signal to bacteria that are also induced save its soul creating many mdr genes and activating gene transfer mechanisms by combining R-plasmids with F’-plasmids. Thus, large conjugative MDR plasmids have 5-15 mdr genes, 6-10 metal resistant genes and two dozen TRA genes for conjugation as well as new protein synthesizing genes like transposases, integrases, topoisomerases, resolvases, restriction endonucleases, DNA ligases and DNA polymerases. Physicians do not know how to cure KPC2 Klebsiella kneumoniae, NDM1 Escherichia coli or MRSA Staphylococcus aureus, MDR Mycobacterium tuberculosis and XDR Acinetobacter baumannii infections. Sadly, once used ampicillin, oxacillin, streptomycin, cefotaxime, azithromycin, tetracycline, ciprofloxacin and chlormphenicol are useless against those bacteria. Our study with Ganga River water of Kolkata indicated that super drugs like imipenem, colistin, tigecycline, amikacin, ceftizidime, vancomycin, levofloxacin and linezolid resistant bacterial species were generated creating a antibiotic dark age even we had thousand antibiotics in the selves.
Keywords
Anti-microbial resistant; MDR plasmids; Beta-lactamases; Drug efflux genes
Introduction
Past 75 years are the golden era of drug development and few thousands penicillin drug derivatives are produced targeting bacterial cell wall peptidoglycan synthesis [1,2]. Dr. Selman A. Waksman discovered over twenty antibiotics from bacteria, actinomycetes and fungi including streptomycin and chloramphenicol that eradicated TB and typhoid in 1950s respectively [3]. New era of biology was begun since 1953 with the discovery of structure of DNA, gene structure, regulation of gene expression and advancement of DNA sequencing, chromosomal structure and RDT work [4-7]. Profound impact was seen in bio-molecules separation by ion-exchange and gel filtration chromatography, glycerol or sucrose gradient ultracentrifugation and HPLC followed by chemical structure analysis by Mass, NMR and FTIR [8,9]. However, last two decades doctors found that drug response curve of many diseases have profoundly changed with increase MIC and a dark site of success RDT work using plasmids pBR322 containing amp and tet genes were anticipated [10-12]. Now every laboratory use dozens of marker genes (neo, amp, tet, aac, cat, pac) derived from drug resistant 5-15 kb R-plasmids. Such plasmids do not transmit mdr genes to other bacteria by conjugation and even such genes have identified in 1940 (sequenced in 1965), it took few decades to generate MDR conjugative plasmids that profoundly were making most bacteria drug resistant. Scientist predicted that antibiotic use by patients, antibiotic contamination from industry, antibiotic use in agricultural land and lives stocks growth, all are contributing to mdr gene creation and spread [13,14].
Results and Discussion
Pubmed and GenBank search suggested that unresponsiveness of many bacteria to drugs like ampicillin, chloramphenicol, streptomycin and tetracycline as early as 1940s forced drug companies to develop new drug derivatives that effective against MDR bacteria [2]. Naturally, semi-synthetic drugs were made without choice to overcome the action of multi-drug resistant genes located in bacterial plasmids that inactivate the antibiotics by different mode of actions [10]. So, journey from 1940-1960, described the isolation of tetracycline, streptomycin, sulfa-drug, ampicillin, amoxicillin, cefoxitin, cefotaxime, erythromycin, nalidixic acid, ciprofloxacin, neomycin, polymyxin, enoxacin, norfloxacin (Figure 1). However, at the almost same time, resistant bacteria to all these antibiotics were developed and the pharmaceutical companies and investors had lost their money (1-5 billion dollar/drug). What happen to investor if a developed drug is good for few years and then drug resistant microbes were appeared when no one would want to prescribe that antibiotic because uncertainty of cure of such infections? In fact, still now R and D Industry is screening new drug every day and also computer-guided graphics design and stimulation of artificial drug-target interactions are accelerating the new drug development. Screening of new drug from fungi was favorable in sense that in soil and water there is a battle between bacteria and fungi and so fungi will produce anti-bacterial to kill bacteria. That type of selection was good having different phylum but what we did introduced actinomycetes derived drug like neomycin and bacteria derived drug like streptomycin from Streptomyces griseus. Then we introduced total synthetic drug against bacteria like ovibactam that was so far good but how long as similar drug sulbactam became useless [15]. What had happened in life of bacteria that all wanted to destroy it by polluting water and consuming antibiotics and as a result bacterium were forced to rearrange its genes to save its own life (Figure 2). In 1960-1980, we produced >1000 tons of antibiotics in industry and 7000 million of global peoples now taken antibiotics almost every month to remove the bacteria from intestine for good health [16]. Does all patients are taking multi-vitamin after each antibiotic dose? Are patients taking probiotics after each antibiotic dose? The answer is simple, in poor nations most doctors do not even prescribe probiotics and multi-vitamins during antibiotic treatment [17]. In Kolkata, I was advised for amoxycillin-cavulinate but sad thing was not advised to take probiotic capsule or multivitamin capsule! So, in my opinion, in most poor and even developing countries we are creating serious health hazards by prescribing antibiotics to all peoples whose metabolism are surely reduced due to lack of coenzymes like FAD, NAD+, COA-SH, THFA, Biotin and vitamin-B12 [18]. The scenario is fatal as we boil most foods destroying vitamins and we do not take raw vegetables and fruits that are great source of vitamins. G-20 leaders and scientists are gathered in Germany recently and have issued Action Plan to reduce superbug horror, but the calamity remains as new therapeutics are not going to develop very soon. Surely, phage therapy clinical trials are ongoing, but phage resistant factors are slowing the FDA approval [17]. Nanotechnology applications are underway to deliver toxic drugs but are very costly. Phytoantibiotics will be considered again but we have cut all plants for crops and a new agricultural land development is necessary but not possible as South Asian countries are highly populated and needs more and more foods which are getting costly day by day. We are educated in science but still we are ill minded as we want to develop poison, hydrogen bomb and nuclear power for mass destruction, not for generating good civilization! How we define human is best creature in this Earth which is still fabricated with many small countries having no scientific development but fight each other like primitive age peoples! We need one nation research and development plans, so all discoveries reach common man at reduced cost. If bacteria can save itself from drugs, then we have to develop new technology to control pathogens. We have new brands of computer, aeroplane, mobile phone, paint, cosmetics, detergent, company with share market, car but we cannot say a name of new antibiotic that available this year against MDR bacteria [19].
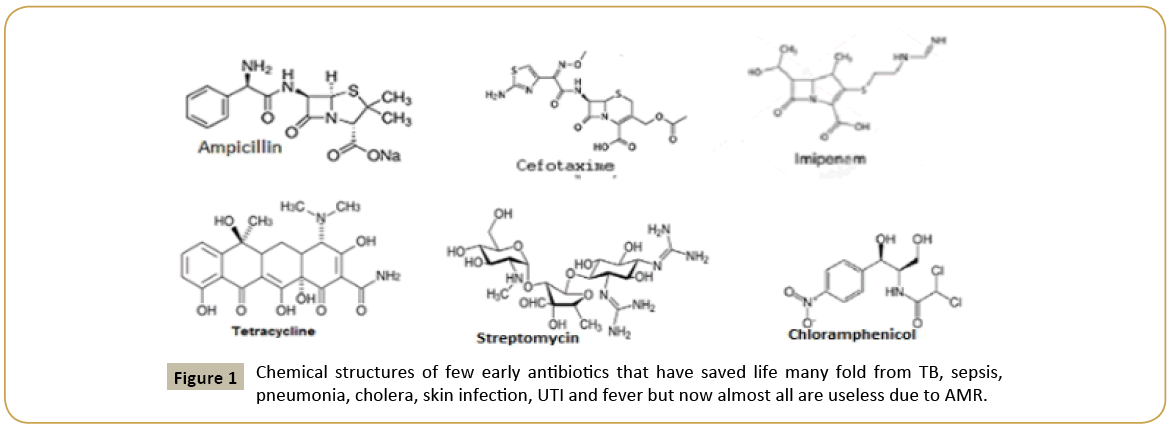
Figure 1: Chemical structures of few early antibiotics that have saved life many fold from TB, sepsis, pneumonia, cholera, skin infection, UTI and fever but now almost all are useless due to AMR.
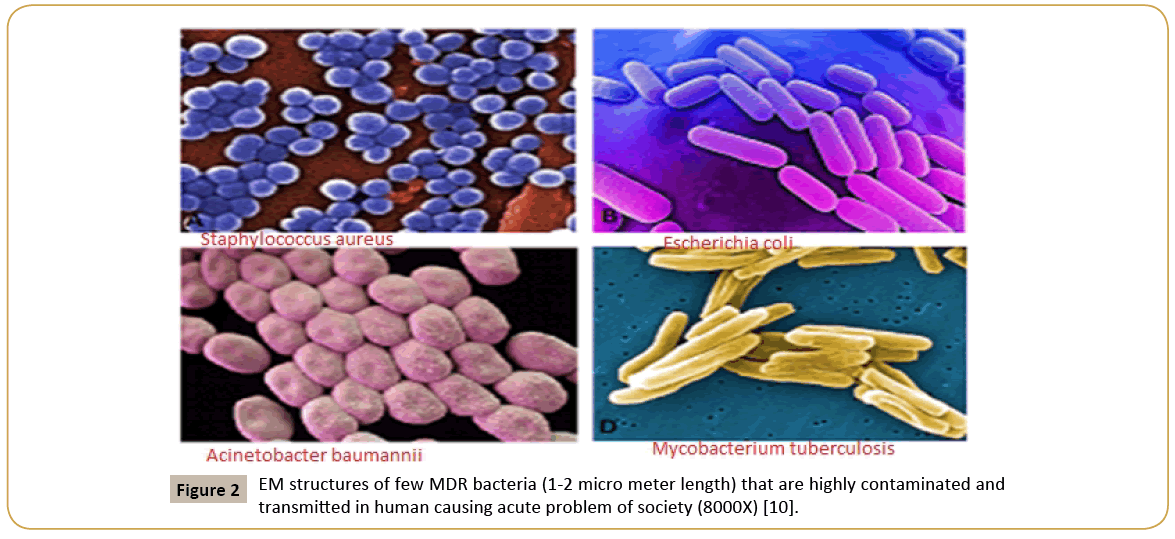
Figure 2: EM structures of few MDR bacteria (1-2 micro meter length) that are highly contaminated and transmitted in human causing acute problem of society (8000X) [10].
It is noteworthy to state the mechanisms that bacteria developed. The first antimicrobial machinery bacteria were created was small drug resistant plasmid or R-plasmids. During the discovery of restriction enzymes and DNA ligase as well as bacterial transformation principle, we developed plasmid pBR322 from un-characterized three resistant plasmids isolated from ampicillin and tetracycline resistant bacteria (1950s). Very soon we made probe to locate mdr genes in many bacteria known as Southern hybridization (1970s) and also able to locate such genes in unknown plasmids by PCR reaction (1980s). Finally, di-deoxy Sanger DNA sequencing principle helped to understand genetic structure of mdr genes and their associated promoter-enhancerrepressor elements that were induced by antibiotics [6,20]. However, bacteria continued its mission to live in presence of antibiotics and created MDR conjugative plasmid combining R-plasmid with F’-plasmid generating a platform for more space for mdr genes in plasmid and more stability and also more active in participation of battle against antibiotics to transfer mdr genes to all resident intestinal bacteria to save symbiosis (Figure 3) (Table 1). So MDR conjugative plasmid now-a-day 50-500 kb and its sequence is known but the functions of 1/4 genes are still unknown [21].
| Major Characteristics of Different MDR Genes in bacteria and Sequence information | ||||
|---|---|---|---|---|
| Genes | Subclass | AA | Protein Id number | Function of the MDR proteins |
| bla | blaTEM | 286 aa | AAB59737, ABI20744 | Degrade ampicillin, amoxycillin |
| blaCTX-M | 291 aa | ABN09669, CAA63262 | Degrade cefotaxime as well | |
| blaKPC | 293 aa | AAG13410, ACB71165 | Degrade cephalosporin and imipenem | |
| blaNDM1 | 270 aa | AGC54622, AFQ31613 | Degrade carbapenems as well | |
| blaOXA1 | 276 aa | AFG30109, CAC82805 | Degrade oxacillin better than ampicillin | |
| blaAMP-C | 382 aa | AAD28044, AHN62490 | Degrade early penicillins but imipenem | |
| tet | tetA | 399 aa | CAA53389, AHC55487 | Kicks out tetracycline |
| tetB | 401 aa | AKJ20239 | Tetracycline efflux isomer | |
| tetC | 396 aa | AGL61405 | Remove tetracycline better | |
| str | strA | 267 aa | AAA26443, ALF35537 | Phosphorylate streptomycin |
| strB | 278 aa | CED95339, AFU91634 | Phosphorylate streptomycin | |
| cat | catB3 | 210 aa | ABP52023, ABC69169 | Acetylate chloramphenicol |
| aac | aacA1 | 185 aa | BAB72153, BAO48019 | Acetylate aminoglycosides |
| aacA4 | 152 aa | AEZ05102, AFV31448 | Acetylate ciprofloxacin | |
| aac(3’) | 286 aa | AAA21890 | Acetylate tobramycin | |
| aad | aadA1 | 263 aa | AAK13440, ABG23480 | Adenylate drugs like streptomycin |
| aadA4 | 263 aa | AAV34365 | Adenylate many drugs | |
| aph | aph(6’) | 266 aa | CAA25854, CAA68516 | Phosphorylate aminoglycosides |
| aph(4’) | 341 aa | CAA24743, CAA27276 | 4’-Phosphorylating isomers | |
| aph(3’) | 264 aa | AAA85506, CAA23892 | 3’-phosphorylating enzyme | |
| RND | acrAB/EF | 1027aa | WP_001132469 | Acriflavin type efflux proteins |
| mexAB | 1045aa | WP_023101049 | Doxorubicin efflux isomer | |
| MFS | norA | 388 aa | BAA14147 | Norfloxacin efflux enzyme |
| mdtE | 385 aa | ABG71588 | Fluoroquinolone efflux isomer | |
| ABC | macB | 664 aa | EMU20415 | Macrolides efflux proteins |
| DrrA | 316 aa | CCK28451 | Drug efflux 12 channels membrane protein | |
| SMR | Mmr | 107 aa | KPU48951 | Drug efflux 4 channels protein |
| emrE | 110 aa | CAA77936 | Drug efflux small isomer | |
| EMR | emrA/B | ~243aa | CAA26964, CAA68299 | Drug efflux protein |
| MCR | mcr-1 | 541aa | WP_049589868 | LipidA-phosphoethanolamine transferase |
| VanR | vanA | 287aa | AKE81063 | Vancomycin resistant proteins |
| DHPS | sul1/2 | ~279aa | AKG90139, BAN19562 | Sulfamethaxozole resistant protein isomers |
Table 1: Multidrug-resistant enzymes with their NCBI GenBank ID and functions contributing AMR [10]. Accession numbers are useful to retrieve primary amino acid sequence and crystal structure of MDR proteins (www.ncbi.nlm.nih.gov/protein).
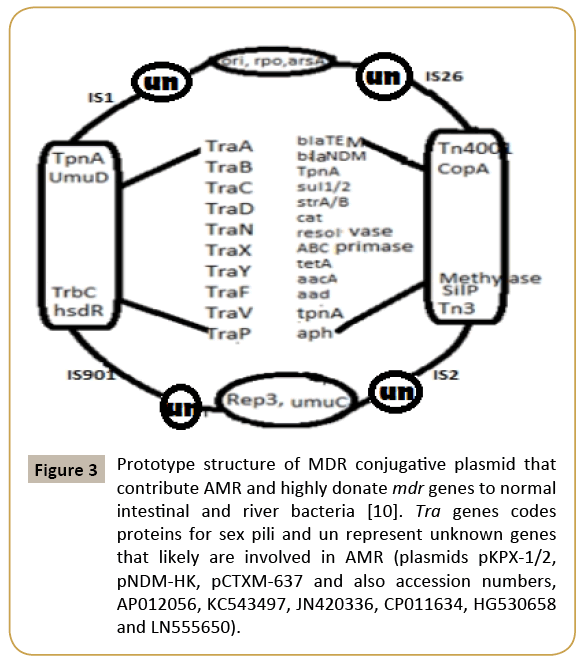
Figure 3: Prototype structure of MDR conjugative plasmid that contribute AMR and highly donate mdr genes to normal intestinal and river bacteria [10]. Tra genes codes proteins for sex pili and un represent unknown genes that likely are involved in AMR (plasmids pKPX-1/2, pNDM-HK, pCTXM-637 and also accession numbers, AP012056, KC543497, JN420336, CP011634, HG530658 and LN555650).
We should not blame only prescription drugs for MDR bacteria generation. Well, large industry like mineral Industry, paint industry, drug industry, paper industry, petroleum industry and excreta from 100 million peoples in many big cities (New York, Mumbai, Kolkata) release tons of chemicals, antibiotics, heavy metals and organic nutrients into river water that are mostly harmful to bacterial central dogma enzymes, but organic nutrients do foster bacterial growth facilitating a huge platform of mutant bacteria creation other than human intestine [22]. So apart from mdr genes (bla, cat, neo, aac, aph, amp); bacteria also created many drug efflux genes by modification of >200 ABC transporter genes which are ATP-driven network of 12- 14 channels membrane proteins involved in nutrient transport across the membrane into cytoplasm [23] (Figure 4 for many mdr genes ). As toxic chemicals and heavy metals are increased in water, it is necessary to develop more gateways to remove toxic chemicals from bacterial cytoplasm. Now few thousand drug efflux genes were sequenced, but their roles in removing antibiotics remains a field of active research. Never-the-less, RND, MFS, MATE types drug efflux genes and proteins have well studied with their capacity to remove penicillins, aminoglycosides and fluroquinolones antibiotics [24]. Perhaps most important frightening fact is such genes are also getting accumulated in MDR conjugative plasmids and chromosome. A 150 kb IncA/C plasmid pMRV150 in Vibrio cholerae 0139 strain was found resistant to common antibiotics, ampicillin, tetracycline, gentamycin and chloramphenicol. A IncC hybrid 165 kb plasmid in Proteus mirabilis was discovered in 2017 with 15 mdr genes including most deadly blaNDM-1 and blaCTX-M-65 and indicated that how severe recombination was facilitated in the human intestine during antibiotics exposure [25]. We detected many mdr genes in Kolkata superbugs (accession numbers: KU898253, KY769876- KY769883) and confirmed by PCR and DNA sequencing (accession number: KY769875 and in preparation) (Figure 5).
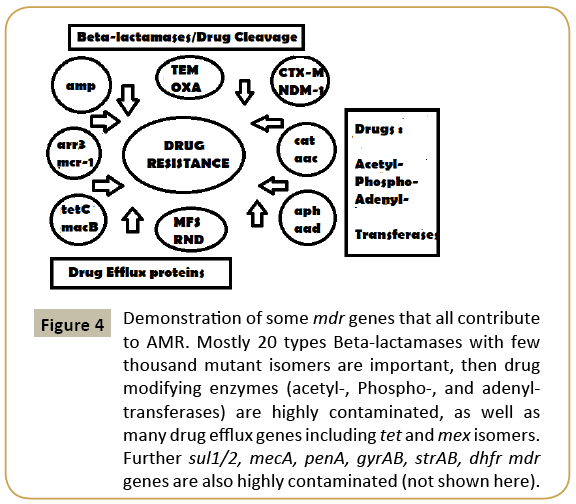
Figure 4: Demonstration of some mdr genes that all contribute to AMR. Mostly 20 types Beta-lactamases with few thousand mutant isomers are important, then drug modifying enzymes (acetyl-, Phospho-, and adenyltransferases) are highly contaminated, as well as many drug efflux genes including tet and mex isomers. Further sul1/2, mecA, penA, gyrAB, strAB, dhfr mdr genes are also highly contaminated (not shown here).
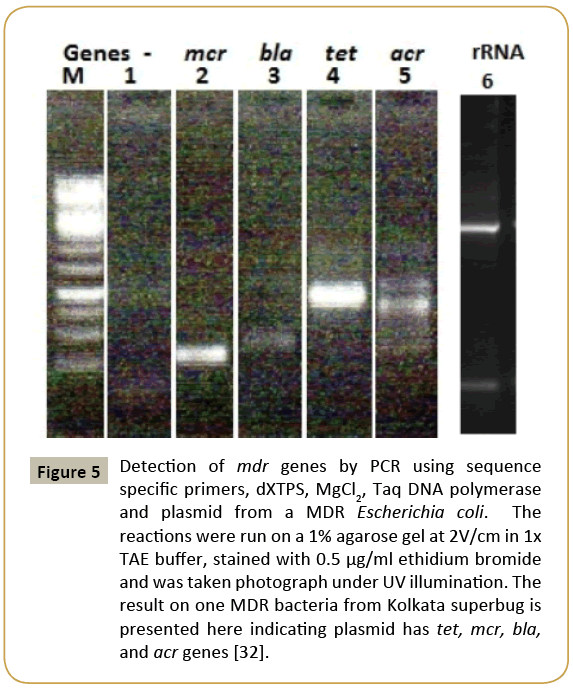
Figure 5: Detection of mdr genes by PCR using sequence specific primers, dXTPS, MgCl2, Taq DNA polymerase and plasmid from a MDR Escherichia coli. The reactions were run on a 1% agarose gel at 2V/cm in 1x TAE buffer, stained with 0.5 μg/ml ethidium bromide and was taken photograph under UV illumination. The result on one MDR bacteria from Kolkata superbug is presented here indicating plasmid has tet, mcr, bla, and acr genes [32].
The adaptation of environment is natural, and bacteria also continued its mission to defect antibiotics by mobilizing mdr genes and drug efflux genes (acrAB, mexCD, norA, macAB) into bacterial chromosome and such genetic islands are well characterized in Escherichia coli, Salmonella typhi, Pseudomonas aeruginosa, Mycobacterium tuberculosis, Helicobacter pylori (cag/T4SS islands) and Acinetobacter baumannii [26-28]. That is not the end, porin membrane proteins are also mutated in such a way that antibiotics receptors are altered, and no drug could enter into bacteria at low drug concentration giving MDR. Further, ribosomal ribonucleic acids (23S, 16S rRNA genes) are gathered few mutations (usually very conserved) as well as ribosomal proteins with alteration of drugs interactions causing MDR. In one word, bacteria have achieved many shrouds against antibiotics and drug companies did not know where to start [29].
Conclusion
WHO warned that if alternative to antibiotics would not discovered soon, very fatal human loss might be occurring in the future? Likely herbal antibiotics research has given priority in India as there is enough medicinal plants and spices available as described in Sanskrit books Charaka Samhita and Veda [30]. However, gene medicines (ribozymes, miRNA, antisense RNA) and DNA nanotechnology have been welcome to stop the horror of MDR bacterial pathogenesis. More sadly, bacteria have acquired promoter induction system by antibiotics and many transcription factor repressors (tetR, mtrR, acrR) have been found in conjugative plasmids [31]. Those TFs induced efflux pumps (mexAB, mtrCDF) and Beta-lactamases (blaNDM1, blaOXA23) could destroy antibiotics and such mechanisms remain elusive. In such a situation, heterogeneous phyto-antibiotics and gene medicines (antisense RNA, ribozyme, Caspase-Cas, miRNA) with different efficient drug delivery modes like DNA nanotechnology are important R and D for new hope. Phage therapy research has forwarded strongly and is waiting for FDA approval [20]. WHO suggested that all countries should follow AMR Action Plan by increasing R and D for new therapeutic intervention as well as by reducing antibiotic use in human, food animal and agricultural land. In other words, we need strong country, one nation platform to keep this Earth as good habitat for human and we have to culture science at every step for our growth. As rate of MDR generation against common antibiotics was high (5% to 10% per year) with 20% to 40% bacteria in water and >95% human pathogens are ampicillin resistant now, it could be assumed that bacteria won the battle against antibiotics and we had entered in an antibiotic dark age as before 1926 [32]. Most importantly, we have houses, schools and shops but no play grounds causing obese, hypertensive and diabetic children with weak immune system that may also contribute a wild spread of tuberculosis (TB) in India. Indian Government has lunched Ganga River Mission but no way we can remove the MDR bacteria and fungi from its water which was consumed raw in religious occasion. In South Asian countries phyto-antibiotic research has given priority [33,34]. But we also need gut microbiota protection awareness and reduced antibiotic use [18].
Acknowledgement
I thank Dr. Bidyut Bandhopadhyay for help during the study and Dr. J. B. Medda for financial help.