Commentary - (2017) Volume 0, Issue 0
Nisreen Al-Moghrabi*, Nujoud Al-Yousef, Maram Al-Showimi and Lamyaa AlGhofaili
Department of Molecular Oncology, King Faisal Specialist Hospital and Research Center, KSA
*Corresponding Author:
Nisreen Al-Moghrabi
Department of Molecular Oncology
King Faisal Specialist Hospital and Research Center
Head of Cancer Epigenetic Section, KSA.
Tel: 966 1 4647272
E-mail: nisreen@kfshrc.edu.sa
Received Date: January 16, 2017; Accepted Date: January 30, 2017; Published Date: February 06, 2017
Citation: Al-Moghrabi N, Al-Yousef N, Al-Showimi M, et al. MicroRNA-126 is Dysregulated in Cancer-Free Females Harboring Methylated BRCA1 Promoter through Up-Regulation of DNMTs. J Clin Epigenet. 2017, 3:1. doi: 10.21767/2472-1158.100037
Copyright: © 2017 Al-Moghrabi N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Background: Numerous microRNAs have been categorized as tumor suppressors that are often repressed by aberrant DNA methylation. Recently, BRCA1 promoter methylation in White Blood Cells (WBC) has been linked to elevated risk of developing breast cancer. In the present study, we aimed to investigative cancerrelated microRNA alterations in WBC harboring methylated BRCA1 promoter. Methods: The Human Cancer miRNA EpiTect Methyl II Signature PCR Array was used to study the methylation status of several cancer-related microRNA sequences. Real time RT-PCR was used to determine the levels of microRNA primary transcripts and DNMTs. The Stem-loop RT-PCR assay was also used to quantify the expression levels of mature microRNAs. Results: Out of 35 miRNAs, miR-126 was found to be hyper-methylated in WBC of breast cancer patients as well as in WBC of cancer-free females who harbored methylated BRCA1 (carriers). The methylation was associated with down regulation of the pre-miR-126 in WBC and lower levels of circulating mature miR-126 in the plasma. Strong positive correlation between the expression levels of both forms of miR-126s was observed in the carriers but not in cancer patients. Importantly, we have found higher mRNA expression of DNMT1, DNMT3a and DNMT3b in WBC of carriers compared to controls. Conclusion: Here, we have demonstrated that miR-126 is dysregulated in WBC of cancer-free females harboring methylated BRCA1 promoter. Importantly, our findings suggest that the up-regulation of DNMTs could be responsible for the aberrant methylation of both miR-126 and BRCA1 promoter in WBC.
Keywords
MicroR-126; Methylation; BRCA1; Breast cancer; DNMTs
Introduction
Epigenetics is known as inherited modifications that influence gene expression without changes in the actual DNA sequence. These modifications include DNA methylation, histone modifications and microRNA expressions. These epigenetic modifications are compelling mechanisms responsible for the inhibition of various genes, including tumor suppressors [1]. DNA methylation is one of the best-studied epigenetic modifications. There are three main DNA methyltransferases (DNMTs) responsible for establishing and maintaining the patterns of DNA methylation in human cells. DNMT3a and DNMT3b are de novo methyltransferases [2,3] while DNMT1 is a maintenance enzyme [4]. The overexpression of these enzymes has been reported to correlate with poor prognosis of several cancers including breast cancer [5-9]. DNA methylation also cooperates with other epigenetic marks such as microRNAs in the complex regulation of gene expression [10].
MicroRNAs (miRs) are short non-coding RNAs that play important roles in regulating gene expression. These RNAs are a novel class of cancer-relevant molecules that are critical for proliferation, differentiation, development, apoptosis and oncogenesis [11]. Several miRs have been characterized as tumor suppressors or oncogenes [12]. The differences in the amount of circulating miRs in the blood plasma of cancer patients compared to that in healthy donor’s offers them as innovative biomarkers for early cancer detection [13]. Several specific miRNAs play important roles in controlling DNA methylation through targeting the DNA methylation machinery. Among the reported miRNAs is miR-126. MiR-126 play crucial roles in several human cancers [14]. It acts as a metastasis suppressing microRNA that is lost in human breast cancer [15]. It has been reported that breast ductal carcinoma patients had lower circulating levels of miR- 126 compared to healthy controls, which correlated with that in the breast tissue [16]. In addition, microRNA-profiling study of the breast carcinoma molecular subtypes has shown that miR- 126 expression levels were able to differentiate malignant from benign breast tumors [17]. These data indicate that miR-126 can act as a tumor suppressor in breast cancer, which could be used as a biomarker for cancer early detection. Interestingly, the circulating levels of miR-126 were found to be significantly higher in the serum of ovarian cancer patients compared to that of controls [18].
MiR-126 is located within the 7th intron of epidermal growth factor like domain 7 (EGFL7), which is itself one of the major targets of miR-126. Both miR-126 and EGFL7 are downregulated in cancer cell lines by hyper-methylation and their expressions can be restored with 5´-aza-2´-deoxycytidine treatment [19]. However, other studies have reported the EGFL7-independent regulation of miR-126 expression suggesting the existence of a separate promoter that derives the expression of miR-126 other than the host gene EGFL7 [20,21].
Similar to germline mutations, DNA methylation is a mechanism for BRCA1 inactivation during breast cancer development [22]. The pathological features of the tumors with BRCA1 promoter methylation are similar to those carrying inherited BRCA1 mutation. Both types occur at an early age and present poor histological differentiation, aneuploidy, ER and PR negativity, as well as similar global gene expression profiles [23,24]. The detection of methylated BRCA1 promoter in DNA from normal tissue has suggested the association of this epigenetic defect with the development of BRCA1-like breast cancer [25]. Several studies have reported the detection of methylated BRCA1 in very young breast cancer patients [26] as well as in healthy females [25,27-31] suggesting the potential use of methylated BRCA1 as a predictor of cancer risk.
Previously, we found a clear link between the aberrant methylation of BRCA1 promoter in WBC and breast cancer-related molecular changes [32]. In the present study we show that miR- 126 expression is dysregulated in WBC harboring methylated BRCA1 promoter through the up-regulation of DNMTs in cancer free females.
Methods
Study samples
This is a continuation of our previous study [32]. We have made use of the available DNA and RNA isolated from WBC and plasma obtained from breast cancer patients and cancer-free females group harboring methylated BRCA1 in their WBC and from cancer-free non-methylated females control group.
Cancer miRNA DNA methylation PCR array
The Human Cancer miRNA EpiTect Methyl II Signature PCR Array (Qiagen) was used to study promoter methylation status of a panel of 22 genes encoding 35 miRNAs. 1 μg of genomic DNA isolated from WBC was used in the array following the manufacturer protocol. An integrated Excel-based template, which is provided by the manufacturer, was used for data analysis. 2.5-fold or greater change relative to controls was determined to be the threshold cut-off point for what is considered a change in gene methylation.
Real-time RT-PCR
WBC RNA was used for reverse transcription using Superscript III (Invitrogen) and random hexamer. PCR was then performed with primer specific for the S2 alternative transcript for the EGFL7 and for the DNMTs as described previously [33,34]. GAPDH mRNA was used as an internal control. Quantitative analysis was performed by real-time PCR with CYBR green using CFX96 real time system (Bio-Rad). The fold change of mRNA expression in breast cancer patients and in carriers relative to the expression in normal controls were calculated based on the threshold cycle (Ct) value using the 2-ΔΔct method.
Stem-loop RT-PCR assay
Mature miR-126 expression was analyzed in plasma. cDNA was synthesized using total RNA, gene-specific stem loop Reverse Transcription primer, and the TaqMan microRNA reverse transcription kit (Applied Biosystems). Quantitative PCR was carried out in a CFX96 real-time system (Bio-Rad) using SsoFast Supermix PCR master mix (Bio-Rad). The fold change of miRNA expression in breast cancer patients and in carriers relative to the expression in normal controls were calculated based on the threshold cycle (Ct) value using the 2-ΔΔct method. Experiments were normalized to small nuclear RNA U6, which was used as internal control.
Statistical analysis
Data are presented as the mean± SEM of replicate experiments. T-test was performed to determine the statistical significance between the different groups for gene expression levels (Patients vs. controls, Carriers vs. controls and Carriers vs. patients). All observed differences were considered to be significant when associated with a P value <0.05.
Results
MiR-126is aberrantly hyper-methylated in WBC harboring methylated BRCA1promoter in both breast cancer patients and carriers
In our previous study [20], we reported the methylated BRCA1 promoter in the WBC of 22 breast cancer patients and 13 cancer free females by screening 155 breast cancer patients and 143 controls, respectively. For simplicity, we termed cancer-free females harboring methylated BRCA1 as “Carriers”. In the present study, we have analyzed the methylation status of cancer related miRNAs, which are known to play important roles in carcinogenesis process. To this end, we used The Human Cancer miRNA EpiTect Methyl II Signature PCR Array, which profiles the promoter methylation status of a panel of 22 genes encoding 35 miRNAs. Based on a cut-off value of +2.5 fold relative to controls, miR-126 was found to be highly methylated in both breast cancer patients and carriers, 4.1 and 3.4 fold, respectively (Table 1). This indicates that WBC harboring methylated BRCA1 promoter exhibit similar alterations in microRNA expression levels in both breast cancer patients and cancer-free females.
| Groups | Fold increase in miR-126 promoter methylation relative to controls | Fold decrease in Pre-miR-126 mRNA expression inside WBC relative to controls |
|---|---|---|
| Patients | 4.1 | 9 (p=0.0004) |
| Carriers | 3.4 | 3 (p=0.003) |
Table 1 Fold changes in methylation and mRNA expression levels in breast cancer cases and carriers.
Pre-miR-126 is down-regulated in WBC of both breast cancer patients and carriers
To investigate the effect of the aberrant hyper-methylation of miR- 126 DNA on the expression levels of pre-miR-126, we analyzed the level of the S2 alternative transcript of the EGFL7 host gene, which corresponds to the precursor of miR-126, in WBC by Real- Time RT-PCR [32]. We have found that the expression of pre miR- 126 was not detected in the majority of the breast cancer patients resulting in significant down regulation in WBC of cancer patients compared to that of controls (p=0.0004). Importantly, we have also found significant down regulation of pre-miR-126 in WBC of carriers compared to controls (p=0.003) (Figure 1; Table 1). This indicates that the aberrant hyper-methylation of miR-126 in WBC associates with a down regulation of pre miR- 126 in both breast cancer patients and carriers.
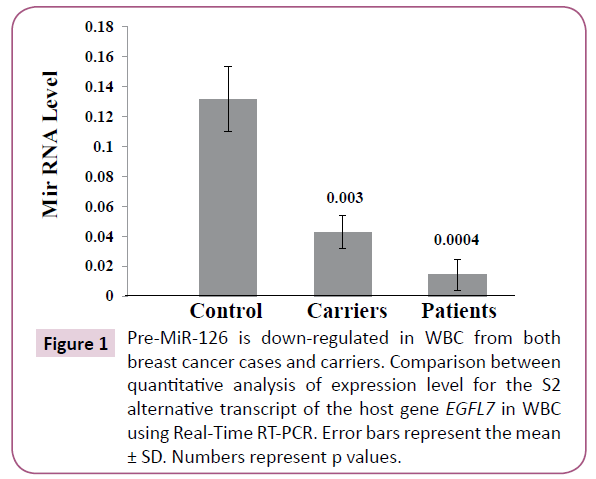
Figure 1: Pre-MiR-126 is down-regulated in WBC from both breast cancer cases and carriers. Comparison between quantitative analysis of expression level for the S2 alternative transcript of the host gene EGFL7 in WBC using Real-Time RT-PCR. Error bars represent the mean ± SD. Numbers represent p values.
Circulating mature miR-126 is under expressed in plasma from both breast cancer patients and carriers
It has been previously shown that miR-126 levels are significantly lower in the sera of breast cancer patients compared with those in normal controls, which correlated with that in tumor tissues [16]. Hence, we sought to investigate whether the decrease in pre-miR-126 in WBC associates with an under expressed mature miR-126 in the plasma. To this end, we made use of the TaqMan miRNA assay with gene-specific stem loop Reverse Transcription primer using total RNA isolated from plasma. We had plasma samples only from four healthy controls, four cancer patients and five carriers available for analysis. Indeed, we have found 7.6 fold decreases in the expression of circulating miR-126 in two of the patients compared to controls (Figure 2A, Table 2); while in the other two patients the expression levels were reduced 2 and 3 folds. Similarly, circulating miR-126 was decreased 2 and 3 fold in plasma from two carriers, CR5 and CR11, respectively, as compared to controls (Figure 2B, Table 3). Importantly, carrier CR21, which have a breast cancer family history (Table 3), showed a significant down regulation in the expression level of circulating miR-126, 16 fold, as compared to controls. Intriguingly, one of the carriers, CR18 with an ovarian cancer family history (Table 3), had 1.6 fold higher levels of circulating miR-126 compared to controls (Figure 2B; Table 3). This indicates that cancer-free females with WBC methylated BRCA1 have dysregulated circulating miR-126 similar to that seen in plasma of breast cancer patients.
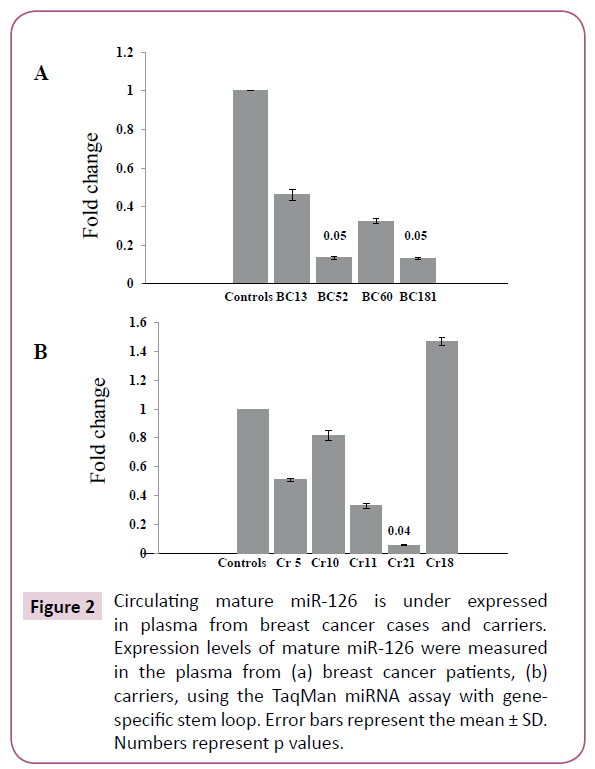
Figure 2: Circulating mature miR-126 is under expressed in plasma from breast cancer cases and carriers. Expression levels of mature miR-126 were measured in the plasma from (a) breast cancer patients, (b) carriers, using the TaqMan miRNA assay with gene specific stem loop. Error bars represent the mean ± SD. Numbers represent p values.
| Patient samples | Fold change | Breast cancer tumor type |
|---|---|---|
| BC 13 | 0.5 | ILC |
| BC 52 | 0.13 | ILC |
| BC 60 | 0.32 | IDC |
| BC 181 | 0.13 | IDC |
ILC: Invasive lobular carcinoma; IDC: Invasive ductal carcinoma
Table 2 Fold change in the expression of circulating miR-126 in the breast cancer patients.
| Carriers samples | Fold change | Age | Cancer family history |
|---|---|---|---|
| CR 5 | 0.5 | 28 | ND |
| CR 10 | 0.8 | 27 | No cancer in the family |
| CR 11 | 0.33 | 26 | ND |
| CR 21 | 0.06 | 21 | Breast cancer (Mother) |
| CR 18 | 1.57 | 41 | Ovarian cancer (aunt, mother side) |
| ND: No Data | |||
Table 3 Fold change in the expression of circulating miR-126 in the carriers.
Mir-126 is regulated at different levels in breast cancer patients and carriers
In order to elucidate the mechanism of miR-126 biogenesis in breast cancer patients and carriers, we sought to assess the relationship between the expression levels of the precursor and mature miR-126 in the two groups. A strong positive correlation was found between the expression levels of the two forms of miR-126 in the carriers with a correlation coefficient of 0.9 (Figure 3A). Interestingly, this was not the case in the breast cancer patients where the mature miR-126 was expressed in the plasma although the pre-miR-126 was not detected in the WBC; hence no correlation was found between the precursor and mature miR-126 (Figure 3B). This indicates that various levels of regulation govern the dysregulation of miR-126 in breast cancer patients and carriers.
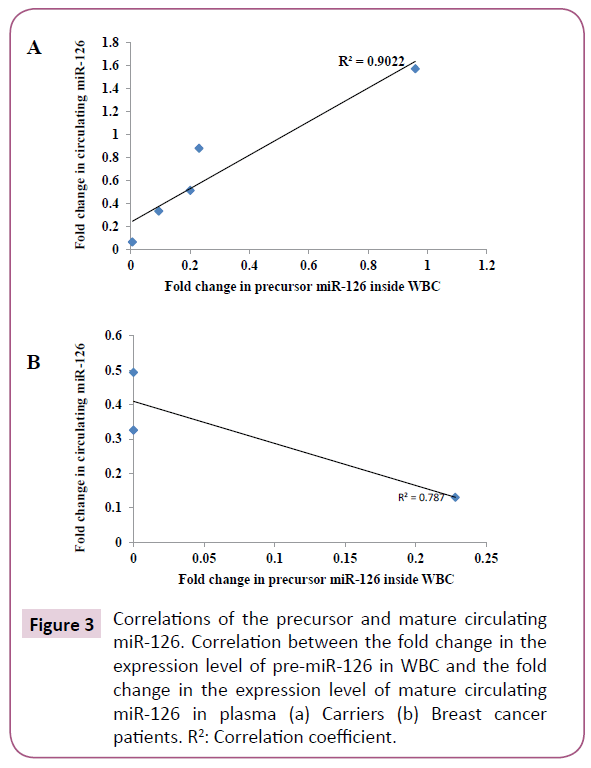
Figure 3: Correlations of the precursor and mature circulating miR-126. Correlation between the fold change in the expression level of pre-miR-126 in WBC and the fold change in the expression level of mature circulating miR-126 in plasma (a) Carriers (b) Breast cancer patients. R2: Correlation coefficient.
DNMT1, DNMT3a and DNMT3b are up-regulated in WBC of carriers
A recent work has shown that the aberrant up-regulation of DNMT1 in esophageal squamous cell carcinomas (ESCC) is responsible for the hyper-methylation of EGFL7, which consequently leads to the down-regulation of miR-126 [34]. Moreover, it has been also reported that the overexpression of DNMT1/3a was correlated with the promoter hyper-methylation and reduced expression of BRCA1 in sporadic breast cancers [9]. Hence, it was tempting to assess the expression levels of all three DNMTs in WBC from the carriers. We used Real-Time RT- PCR utilizing primers for DNMT1, DNMT3a and DNMT3b. Intriguingly, we have found 1.7 fold (p=0.04) increase in the mRNA expression level of DNMT1 (Figure 4A) and 2-fold increase for both DNMT3a (p=0.08) and DNMT3b (p=0.1) (Figure 4B and 4C) in carriers compared to controls. This suggests that the up-regulation of the DNMTs could be responsible for the aberrant methylation of miR- 126 as well as BRCA1 in WBC of carriers.
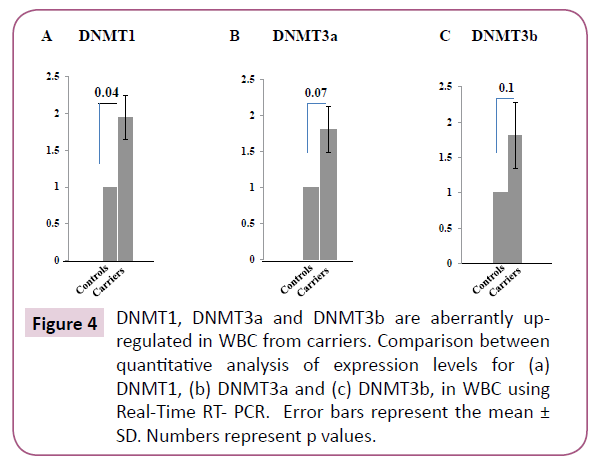
Figure 4: DNMT1, DNMT3a and DNMT3b are aberrantly up regulated in WBC from carriers. Comparison between quantitative analysis of expression levels for (a) DNMT1, (b) DNMT3a and (c) DNMT3b, in WBC using Real-Time RT- PCR. Error bars represent the mean ± SD. Numbers represent p values.
Discussion
The abnormal expression levels of mature and precursor miRNA transcripts are considered to dysregulate miRNA expression that plays an essential role in cancer initiation and progression. In the present study, we report the aberrant expression of the precursor and the mature miR-126 in WBC and plasma, respectively, in both breast cancer patients and carriers.
MicroRNA-126 has been suggested to serve as a non-invasive biomarker for breast cancer. Patients with breast cancer had lower circulating levels of miR-126 when compared to normal controls [17]. In addition, it has been shown that the down-regulation of miR-126 [35] is associated with triple-negative breast cancers. Here, we have found that miR-126 is highly methylated in WBC of both breast cancer patients and carriers. We also found that this methylation to be associated with significant decrease in pre miR- 126 expression in WBC from both groups.
Previously [32] we have found significant association between the presence of BRCA1 promoter methylation in WBC and the cancer incidence in the families of the carriers. Interestingly, in the present study, we have also found that the dysregulation of miR-126 is associated with the cancer family history of the carriers. The carrier CR21, which have a breast cancer family history, showed the lowest expression levels of the pre-miR-126 in WBC and circulating mature miR-126 in the plasma. Moreover, the fact that carrier CR18, with an ovarian cancer family history, showed an increase in the relative expression of circulating miR- 126 suggest a link between BRCA1 methylation and ovarian cancer predisposition; as circulating miR-126 has been found to be up-regulated in the serum from ovarian cancer [19].
The positive correlation between WBC pre-miR-126 and mature plasma circulating miR-126 levels in the carriers suggest that miR-126 is regulated at the transcriptional level in these individuals. However, the lack of this correlation in the breast cancer patients also suggests that during the carcinogenesis process miR-126 is post-transcriptionally regulated. Indeed, it has been previously shown that pri-miRNA and mature expression is positively correlated in normal tissue but not in breast tumors [36]. These results suggested a multistep model for miRNA dysregulation. The fact that, in the carriers, we have found a more significant decrease in the expression level of the pre-miR-126 than the decrease in the mature form might be an indication for the multistep dysregulation of this miRNA in those individuals.
The up-regulation of DNMT1 in WBC of carriers harboring hyper-methylated miR-126 is in concordance with recent work showing the existence of a “DNMT1-miR-126 epigenetic circuit” in ESCC [27,38]. The aberrant up-regulation of DNMT1 in ESCC is found to be responsible for the hyper-methylation of EGFL7, which consequently leads to the down-regulation of miR- 126. Additionally, the up-regulation of DNMT3a and DNMT3b together with the hyper-methylation of BRCA1 in the carriers are also in agreement with the study showing a correlation of the over expression of DNMT1/3a with promoter hyper-methylation of BRCA1 in sporadic breast cancer. All in all, this may implies that the aberrant up-regulation of the DNMTs mRNA in WBC is responsible for the hyper-methylation of miR-126 as well as BRCA1 in WBC from carriers.
In conclusions, we have clearly demonstrated that miR-126 is dysregulated in cancer-free women harboring methylated BRCA1 promoter in their WBC. The up-regulation of the methylation machinery, DNMTs, could be responsible for the aberrant methylation of miR-126 as well as the methylation of BRCA1 promoter in WBC from carriers. All of the three DNMTs, DNMT1, DNMT3a and DNMT3b, are known to be highly up-regulated in breast tumors [39]. Our findings provide additional support for the presence of a strong link between the aberrant BRCA1 promoter methylation in WBC and cancer-related molecular changes.
Acknowledgment
We are grateful to the patients and controls who participated in this stud. We would like to acknowledge Dr. Abdelilah Aboussekhra and Dr. Bedri Karakas for critically revising the article. We would like to thank the research center administration for its continuous encouragement and support.