Review Article - (2022) Volume 8, Issue 1
Mistranslation of Genetic Codon and Their Controlling Mechanism in Cell
Mohsin Shad1*,
Ayesha Malik2,
Momina Javid2,
Salah ud Din2,
Sana Shakoor2,
Kanza Sadiq2,
Muhammad Ayoub1,
Muhammad Usman3,
Muhammad Sajjad1,
Muhammad Akhtar Ali1 and
Abdul Qayyum Rao2
1School of Biological Sciences, University of the Punjab, Pakistan
2Centre of Excellence in Molecular Biology, University of The Punjab, Pakistan
3Department of Plant Pathology, University of Agriculture, Pakistan
*Correspondence:
Mohsin Shad, School of Biological Sciences, University of the Punjab,
Pakistan,
Email:
Received: 28-Dec-2021, Manuscript No. IPBMBJ-21-11335;
Editor assigned: 30-Dec-2021, Pre QC No. IPBMBJ-21-11335 (PQ);
Reviewed: 13-Jan-2022, QC No. IPBMBJ-21-11335;
Revised: 18-Jan-2022, Manuscript No. IPBMBJ-21-11335 (R);
Published:
31-Jan-2022, DOI: 10.36648/2471-8084.22.8.51
Abstract
Mistranslation is the incorporation of errors during translation, which may lead to the mis-incorporation in the
polypeptide chain by an altered amino acid that is different from the one being translated by the normal gene.
Mistranslation was previously known as the root of disease, but current studies suggest that some organisms
have adapted numerous pathways to tolerate the translation error such as fitness can be increased by mistranslation
through eradicating deleterious mutations. It can also protect bacteria against oxidative stress. A cell can
control mistranslation by different controlling mechanisms. This study discusses different controlling mechanisms
used by a cell to control the misincorporation of amino acids. Mistranslation can be controlled by aminoacyl-tRNA
synthetase (aaRSs) editing function, discrimination against mis-acylated aminoacyl tRNA (aa-tRNA) by EF-Tu,
and by giving up the right substrates for protein synthesis. By manipulating the controlling mechanism of mistranslation,
it can be utilized as a tool to produce novel biopharmaceuticals, for curing disease and to enhance
the cell potential against environmental stresses.
Introduction
The maintenance of a specific proteome can be started with
a high level of translation control which can be highlighted by
the comprehensiveness of the genetic codon. However, a variation
between universal genetic codons is especially frequent.
The first deviations from the genetic code were recognized in
nonsense and frame shifting mutations, and analyses of yeast
and human mitochondrial genomes. Standard genetic code can
be changed naturally to cause uncertain decoding, codon reassignments,
and natural genetic code elongation, such as UAG
and UGA stop codons can be decoded with pyro-lysine and
selenocysteine [1].
Mistranslation is the incorporation of fault during protein synthesis which can be lead to misinforming of an amino acid
that is diverse from being translated by the gene [2]. From all
domains of life genetic information must flow from codon to
an mRNA and then polypeptide chain [3]. The hereditary sequence
is transcribed into mRNA and then translated into protein-
peptide are the mechanisms that can abide some errors in
these processes. Inaccurate mRNA codon can be produced by
incorrectly transcribed DNA sequences; as a result, an unrelated
polypeptide chain can be produced by decoding mRNA at
the ribosome [4]. Misfolding and shortening of the protein can
be done by a fault in central dogma processes which leads to
loss of the protein function at a molecular and cellular level by
deleterious the essential residue [5,6].
Genetic information flow from DNA sequence to protein sequence is the inexact mechanism. Messenger RNA is transcribed
from DNA template and then translated into protein sequence
at the ribosome involve a minute level of errors that produced
a low level of variable proteins sequence [6]. Aminoacyl-tRNA
synthetase has a high level of quality control mechanism that
only binds with amino acid-associated tRNA to form aminoacyl-
tRNA which is utilized at ribosome for polypeptide synthesis
[7]. Impaired aminoacyl-tRNAs are produced by the error in
aminoacyl-tRNAs synthetase function that causes inaccurate
insertion of amino acid which is not specified by genetic codon
through a phenomenon called ‘’mistranslation’’ [2,8]. The loss
of protein structural and functional integrity can be linked with
the departure from genetic codons that cause phenotypic defects
and disease [9].
The expressions of genetic information into functional proteins
tolerate some error during these processes. Mistranslation was
previously familiar as the root of disease [10,11], but current
research showed that certain domains from all life have adapted
numerous pathways to tolerate the translation error [12].
However the major cause for these faults is not identified but
evidence indicates that the higher rate of mistranslation has a
significant role in the protein structure and function [13,14].
The mistranslation error rate can be regulated and differ by
environmental situations and organism diversities [9]. Protein
translation errors have the adaptive role for understanding by
experiment on the wild type of organism and unbiased investigation.
Importance of Mistranslation
As reported earlier, mistranslation occurs due to the faulty incorporation
of amino acids that are dissimilar from the ones
coded by the factors. Mistranslation has a big importance in the
field of science as it can prove to be worthy because of the effect
caused by it.
Mistranslation can be Increased Fitness Levels
by Removing Deleterious Mutations
Variance is the basic material for Darwinian evolution, which is
produced by mutation in the genetic material during replication
and gene expression. The best source for such error is caused by
DNA mutations but mainly errors involves during transcription
[15-17], tRNA, amino-acylation [18], and expression of mRNA
into protein. Protein synthesis is the major possessing error in
the central dogma process [15,19,20]. In E. coli, the rate of DNA
point mutation could occur at the rate of 104 by the mistranslation
rate of 10−5–10−3 per codon [15]. Phenotypic mutations
‘are caused by an error that the ribosome possesses during
mistranslation [15,21]. They may involve frameshift mutation
or missense mutations [20]. Even though they are not as permanent
as DNA mutations, phenotypic mutations have might
an impact on evolutionary processes through their erect abundance.
Mistranslation is combined with natural selection may
have a critical role in the reduction of a protein’s propensity to
misfold because the function of protein might be stabilized by
natural selection [15,20,22].
Mistranslation can speed up adaptive evolution, as a valuable
phenotypic mutation could create a high fitness protein from
a low fitness genotypic intermediate. For instance, during the selection of antibiotic resistance, such beneficial phenotypic
mutations can increase the activity of an enzyme that deactivates
antibiotics. During the appropriate robust selection, even
a few high fitness proteins might increase the survival of the
population and by the time genetic mutations make the adaptive
change permanent, high fitness proteins increase the population
time [15,23]. It is unidentified if such ‘stepping stone’
proteins occur in the formation of new protein functions by
amino acid changing mutations, while they play role in the
production of new protein localization signals via read through
mutations [24]. Furthermore, mistranslation can help adaptive
evolution by exacerbating the deleterious effects of mutations.
As mistranslation enhance the effectiveness of purifying selection
[22], it might support removing deleterious mutations from
standing genetic variation, consequently rising to mean population
fitness.
To inspect how protein mistranslation may influence the evolution
of a novel protein function, scientists conducted laboratory
based evolution experiments on the antibiotic resistance
enzyme TEM-1 β-lactamase. Wild type (‘ancestral’) TEM-1 inactivates
β-lactam antibiotics. It is tremendously active against
penicillin, such as ampicillin. While it demonstrates insignificant
activity on cephalosporins, like cefotaxime, it could develop
high activity against them [15,25]. They tentatively created
TEM-1 to action on cefotaxime in host cells of E. coli subject to
either typical or raised rates of mistranslation and categorized
the advanced populaces both genotypically and phenotypically
employing high-throughput sequencing. We validate that TEM-
1 populaces progressive in mistake strains inclined towards error
are more genotypically diverse and pass on the advantage
of fitness to their host. This benefit is linked to the amplified
riddance of mutations that can surge mean population fitness
under directional selection for a new antibiotic resistance phenotype.
When stabilizing selection occurs, mistranslation may
diminish rates of mistranslation or increment vagueness to
mistranslation [22,26]. In what way impacts of mistranslation
interface during directional and stabilizing selection [15] process
remain a vital inquiry for upcoming research. For instance,
mistranslation may not just have been uncontrolled in initial life
forms [15] yet is commonly more successive even at present
than hereditary changes, it stays a conceivably vital power in
the advancement of present-day proteins. Our perceptions increase
the likelihood that mistranslation might infrequently accelerate
versatile advancement by purging deleterious changes.
Protein Mistranslation Protects Bacteria against
Oxidative Stress
As of late, it has been examined that protein mistranslation
can spare microscopic organisms from oxidative stress. Protein
mistranslation is versatile and useful under oxidative stress conditions
in E. coli. Mistranslated proteins in E. coli activate the
general stress reaction and are also the cause of adaptations
against serious oxidative stress conditions. Mistranslation most
of the time happens in normal E. coli isolates, and is initiated via
aminoglycosides, carbon starvation, viral disease, and oxidative
stress [27-30].
Mistranslation against Diseases
As of late, it was found that the NPA β-amino methyl-alanine (BMAA) is mistranslated in human tissues at serine (Ser), codons
[4,31]. The level of this mistranslation radically diminished
by serine supplementation in the media, recommending
that the source of this mistranslation might be the competition
of non-cognate BMAA with cognate Ser for seryl-tRNA
synthetase. There is an expanded risk for neurodegenerative
disorders with ingestion of BMAA, for example, Parkinsonism,
amyotrophic parallel sclerosis, and Alzheimer’s disease [4,32].
It is an enticing view to utilize nutritional supplementation with
similar amino acids as a treatment for maladies, for example,
as proteinogenic amino acids production is inexpensive, they
are widely accessible over the counter, and are ordinary metabolites
for humans. It’s possible that modifying their selectivity
of aaRSs by chemicals, initiating changes in amino acid
pools, or planned NPA action might be utilized as a part of various
strategies to battle numerous maladies with minor side
defects for the patient.
Controlling Mechanism of Mistranslations
Protein synthesis has achieved a middle of the road level of
loyalty with an error rate of 1 out of 10,000 [19]. There are
various repetitive pathways to check precision and accuracy
in protein translation. The most imperative include the basic
step of tRNA aminoacylation, which joins hereditary data (as
the tRNA anticodon) with protein data (as the attached amino
acids). Some aaRSs have low error rates inherently, while
other aaRSs have an ability that identifies when the inaccurate
amino acid is ligated to the tRNA and hydrolyses this bond to
take into consideration another round of ligation [2]. Several
other quality control systems exist to shield translational mechanism:
aminoacyl-tRNA synthetases (aaRSs) proofreading, mismatch
tRNA deacylation, EF-Tu discrimination against mis-acylated
aminoacyl tRNA (aa-tRNA), legitimate codon-anticodon
arrangement in the ribosomal A site, Watson and Crick base pairing of codons and anti-codons at the ribosome, and last
but not the least ribosomal proofreading mechanism that was
discovered recently [33,34].
The timely synthesis of RNAs can be done by regulating gene
expression. Similarly, the proteins that can meet the needs of
cells e.g., changing developmental and environmental states,
can also be produced. In gene expression, protein synthesis
is the final step. It is the step in which the data which is encoded
as mRNA is translated into the final product in the form
of chains of amino acids which make up the proteins. mRNA
which was synthesized should be completely translated so that
the cell can take benefit from the gene expression [2,9]. Moreover,
translation takes place at a biologically appropriate rate,
a cell that splits every 20 minutes gets little advantage from accurate
protein synthesis that takes 1 hour to finish. Like higher
precision comes with a price like other natural procedures, and
through translation rate of protein formation and the speed of
mistranslation should be precisely adjusted [2].
Aminoacyl-tRNA Synthetase (aaRSs) Editing
Function
Aminoacyl-tRNA synthesis is the initial step of translational
mechanism at which error can happen at a significant rate and
the part of the aaRSs and their editing function in keeping up
translational accuracy is an area of intemperate study [23-28].
A precise choice of amino acid and its binding to the proper
tRNA is needed for Aminoacyl-tRNA synthesis. The binding of
close related amino acids is prevented by aaRSs through various
quality control components of the two step aminoacylation
reaction [33,34]. By especially choosing related sets of amino
acids and tRNAs during discrimination of close and non-related
molecules, aaRSs give a critical initial phase in quality control
during translation (Figure 1).
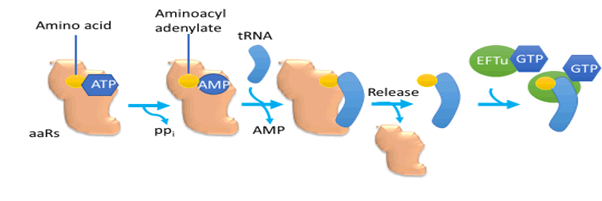
Figure 1: Quality control step during the formation of a cognate amino acid-tRNA pair
Quality control mechanism applied firstly on aminoacyl t-RNA
synthetase active site, at this site amino acids were selected.
However, twenty naturally occurring amino acids do not show
adequate ranges of functional groups functional groups in
some may vary by only one (OH- or CH4+ groups) that enables
aminoacyl tRNA synthetases to distinguish with accuracy level
consistent to a rate of errors ordinarily related with translation,
therefore errors may sometimes happen. Such as valine contrast
by just one CH4+ group from isoleucine [35]. Escherichia
coli and Cerevisiae demonstrated a connection of inaccurate amino acids now and again that can be corrected through editing
before transfer or editing after transfer [33].
Editing before a transfer can happen through different mechanisms
ranging from non-cognate and labile amino-acyl adenylate
class hydrolysis and the non-cognate aa release both are
promoted and this editing before transfer may or may not depend
on t-RNA [36,37]. Editing after the transfer of amino-acyl
t-RNA (non-cognate) needs a CCA tail at 3’ transfer RNA end,
that’s connected to aa. When it moves to the editing site from
synthesis one, the site where hydrolysis of aa-RNA ester link age occurs. This whole process can happen through direct CCA
translocation in cis ones while in trans after aminoacylation
release of the active site of t-RNA occur which then leads to
rebinding of amino-acyl t-RNA to aminoacyl t-RNA synthetase
at the site of editing. Some of the aminoacyl tRNA synthetases
likewise compete with EF-Tu (elongation factor Tu) successfully
for aminoacyl-transfer RNAs, consequently permitting re-examine
of transfer RNA which was mis-acylated and this why got away from the first run of this editing mechanism. That extra
layer of aminoacyl transfer RNA synthetase quality control
builds decoding accuracy of more than ten times in a protein
synthesis system derived from E. coli [38,39]. Editing action after
the transfer isn’t restricted to aminoacyl transfer RNA synthetase
and includes many trans-acting elements in the trans
editing procedure including d-aminoacyl tRNA deacylated [40]
(Figure 2).
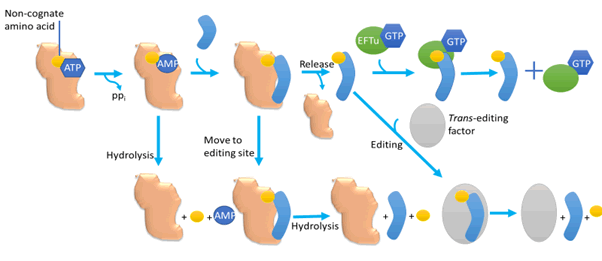
Figure 2: Quality control steps during the formation of a non-cognate aminoacyl-tRNA
Although the inherent aaRS sites for editing are profoundly
saved amongst species the trans-acting elements are significantly
more diversified in their structure and distribution,
recommending that during evolution specific mechanisms for
quality control may have been chosen for more than one time.
Such as for the viability of cells, d-aminoacyl-tRNA deacylated
are required 3 disconnected groups of transfer RNA deacylated
were recognized such as: in microorganism mostly and in
eukaryotes d-Tyr-t-RNA deacylated 1 (DTD1), in cyanobacteriaDTD3
and last class named as DTD2 is found mostly in plants
and in archaea too [41].
Right Substrate Delivery for Protein Synthesis
After alteration and resembling by aminoacyl transfer RNA
synthetase and added trans-acting factors, a ternary complex
formed by aminoacyl-tRNAs with eFTu–GTP this complex then
binds to the ribosome subsequently [39]. The transfer RNA
and aa moieties accomplish contributions related to thermodynamics
during elongation factor EF-Tu binding than in result
all aminoacyl-transfer RNAs which are cognate binds with the same affinity but aminoacyl-tRNA that are non-cognate binds
with affinities that may vary by more than 700 fold [42].
Although eFTu discrimination is not such specific as aminoacyl
transfer RNA synthetase editing, it may accommodate added
band to the quality control mechanism of translation which can
alleviate translation accuracy. Consecutive checkpoints in this
mechanism ensure that the aminoacyl-tRNAs are constructed
with no error and handed in the aboriginal form to elongation
factor Tu and to ribosomes then. Current affirmation proposes
that in higher eukaryotes the whole procedure occurs in a
single complex which is linked with polysomal ribosomes. Numerous
aminoacyl transfer RNA synthetases are well-arranged
in multi synthetase complexes of very high molecular weight
these high weight molecular complexes also include elongation
factors of translation [43]. Now, these multi synthetase
complexes by channeling aminoacyl-tRNAs from the aminoacyl
transfer RNA Synthetase to elongation factors and then towards
the ribosome are contributing toward efficient protein
synthesis mechanism [44] (Figure 3).
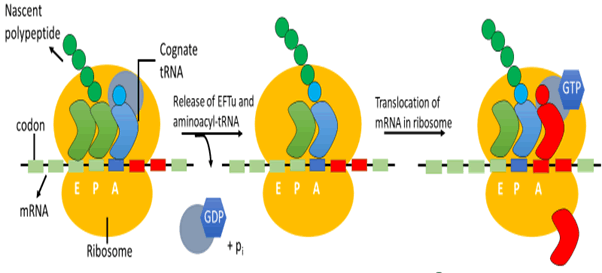
Figure 3: Formation of a polypeptide in the ribosome
Ribosomal Protein Synthesis
All aminoacyl transfer RNAs apart from initiator aminoacyl tRNAs
binds to A site while the latter binds to A site. Hence, the
alpha-amino group of aminoacyl transfer RNAs present at the
A site reacts with the carbon of the carbonyl group of aminoacyl
transfer RNA present at the P site, then a bond is formed
known as a peptide bond. Ribosomes are the place where the
Synthesis of proteins occurs (Figure 4). In the bacilli and archaea domain, this ample ribonucleoprotein is consists of the
30S and 50S subunit, which forms 70S ribosome, and in the eukaryote
domain, it consists of the 40S and 60S subunit, which
results in 80S ribosome [45].
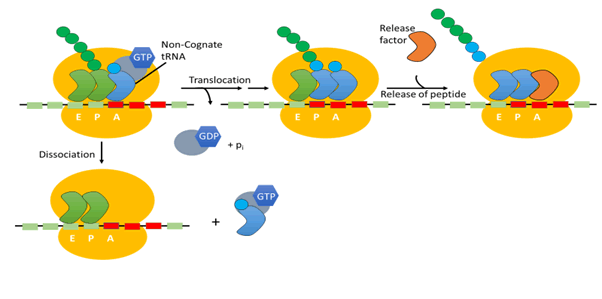
Figure 4: Quality control in the ribosome
Ribosomes accommodate three transfer-RNA bindings’ sites:
the acceptor (A), the peptidyl (P), and the exit (E) all of these
sites participate in translational quality control mechanisms
[46].
Transfer RNA that is 1 aa long known as Peptidyl transfer RNA is
present on A site while the one that is deacylated, deacylated
transfer-RNA is present in P site left to A site. Then 1 codon
is translocated by ribosome alone the messenger RNA, that in
results move the newly deacylated one into E site known as
exit site, peptidyl one to P (peptide) site and A site known as
acceptor site is now free to acquire a new incoming amino-acyl
t-RNA. The last step of accurate protein synthesis after related
aminoacyl transfer RNA transportation to the ribosome acceptor
(A) site, is exact m-RNA and t-RNA anti-codon pairing on the
peptidyl (P) site a small decoding center On a small ribosomal
subunit [47,48].
Proper and improper interactions between codons and anti-codons
can be differentiated by ribosomes, but ribosomes cannot
find out transfer RNAs that are is acylated. substantial post-transcriptional
changes are present in transfer RNA anticodons that
during identification of codons can restrict or either extend the
wobble effect and hence play an important role in the accuracy
and speed of translation [49]. These 7 modifications either
provide structural diversities and this codon anti-codon pairing
is not all alone sufficient for an account of decoding accuracy
which is also accomplished by the ribosomes [2].
It is predicted that if the decoding specificity is gained completely
from base pairings of codons and anticodons, then about
half of the codons would have been translated with about less
than 99 percent perfection, which is far off the error rate of 1 in
103 to 105 codons [50]. There are two step checks for a pairing
of codon and anticodons for decoding at the A (acceptor) site,
first at the initial selection and the 2nd one at proof reading.
At early selection, codon-anticodon pairing is examined, and
the ternary complex is made up of amino-acyl-transfer RNA,
elongation factor T and GTP bind to the ribosome. Remnants of Ribosome at A site of small (30s) subunit monitors the codon-
anticodon helix geometry and this geometry contributes
a lot towards ternary complex conformational changes which
enhance forward reaction rate of GTP-hydrolysis by the elongation
factor Tu [51]. In contrast to this if in codon-anticodon
helix mismatch occurs. it does not allow conformational changes
to occur that are necessary for fast GTP hydrolysis, hence enhancing
the chances that this ternary complex will be dissociated.
After this first step of selection hydrolysis of GTP occurred
and aminoacyl transfer RNA was released by elongation factor
Tu, then this aminoacyl-tRNA either enters Large (the 50S) ribosomal
subunit A site or was rejected from the ribosome. If a
mismatch occurs in codon-anticodon pairing aminoacyl transfer
RNA is probable to be ruled out mostly. This step of translation
quality control provides another independent selection
step. After the successful binding of amino-acyl transfer RNA at
the A-site in the ribosome and contributes towards bond (peptide
bond) formation, this newly synthesized peptidyl-transfer
RNA is transferred to the P site from the A site.
Ribosomes continuously examine at this stage of translation
that codon-anticodon helix does not allow those conformational
changes that are necessary for quick GTP hydrolysis, which
results in an increased probability of ternary complex separation.
GTP is hydrolyzed after this determination step and elongation
factor Tu discharge the amino-acyl transfer RNA. This
aminoacyl transfer RNA either goes to a larger ribosomal subunit
(the 50S) A site or may be rejected by the ribosome, and it
is rejected when a mismatch occurs between codon-anticodon
interaction. Ribosome at this step of translation check for those
mistakes in a codon-anticodons pairing which by chance are
missed at amino-acyl transfer RNA charging steps and in proofreading.
If the codon-anticodon mismatch is available at the P
site it means loss of specificity of the ribosome at A site which results in an intensification of mistakes. This then untimely result
in the premature end of the elongation step and peptides
are released at an accelerated rate.
The ribosome not only checks codon-anticodon associations in
both A and P sites, nevertheless it keeps up the right perusing
outline in the messenger RNA to counteract frameshift mutations,
which additionally prompt mistranslation and untimely
results in termination. Even though the molecular system of
maintenance of the reading frame is not crystal clear, both
the P and E locales of ribosomes appear to be included in this
mechanism. The exit (E) site is particularly vital in the reading-
frame maintenance, as both the codon-anticodon connection
strength and its occupancy in the exit (E) site specifically
add to frame-shift mutations (Table 1).
Table 1: Substrate discrimination mechanism in different steps of translation
| Step in translation |
Substrate discriminated |
Mechanism of discrimination |
Discrimination factor |
References studies |
| Aminoacyl-tRNA synthesis |
Amino acid |
Amino acid recognition |
200–10,000 |
29 |
| Aminoacyl-tRNA synthesis |
tRNA |
Specific binding of tRNA to the aaRS by nucleic acid recognition |
>10,000 |
22 |
| aaRS editing and aaRS resampling |
Aminoacyl-tRNA |
Amino acid recognition |
>10 |
31 |
| EF-Tu binding |
Aminoacyl-tRNA |
Thermodynamic compensation |
Unknown |
46 |
| Initial aminoacyl-tRNA selection |
tRNA |
A site codon-anticodon pairing |
30–60*‡ |
34 |
| Proofreading and accommodation of aminoacyl-tRNA |
tRNA |
A site codon-anticodon pairing |
20–100 |
46 |
| Post-peptide bond formation quality control |
Peptidyl-tRNA |
P site codon-anticodon interaction |
>10 |
48 |
| *By experimentally determined rates of catalysis for GTP hydrolysis based on cognate substrates or near-cognate substrates; ‡Presuming equal concentration of cognate substrates & near-cognate substrates. aaRS, aminoacyl |
Discussion
Need of Quality Control Mechanisms
Quite a bit of our insight into interpretation quality control
mechanisms depend on in vitro examines that portray observation
at every specific or advanced step of translation. Although
this approach has given an abundance of nuclear determination information, extreme little is thought that in what
way, where and at what time quality control mechanisms have
a part in the cell. It is frequently expected that foremost to
keep up the most elevated conceivable level of quality control
and the least interpretation mistake rate. Few microorganisms
Surely hold stringent quality control components aimed at the
synthesis of amino-acyl transfer-RNA. Such as, E. coli phenylalanine
separates or discriminates phenylalanine over Tyrosine by
a segregation factor of greater than 8,000 to 1 (that means, it
mis-incorporates a Tyrosine rather than Phenylalanine just one
time per 8,000 duplicates of transfer R-NA Phenylalanine) and
furthermore keeps up altering action against Tyrosine-transfer
RNA Phenylalanine. Similarly, E. coli and Haemophilus influenzae
Proline discriminates Alanine by greater than 3,000
to 1, contains cis altering domain and their editing exercises
were additionally expanded by the nearness of the trans-acting
cys-tRNA Proline deacylated YbaK [52].
Quality control mechanisms are administered for viability despite
their extensive conservation [2,53]. Determination of the
selective pressures that lead to the extensive conservation of
the quality control mechanisms controlled by aaRSs requires
the recognition of all the conditions under which quality control
mechanisms are needed for the ideal growth of cells.
Lacking the quality control mechanisms not only lead to the
wastage of nutrients when mistranslation produces inactive or
terminated polypeptides, but it also causes the aggregation of
misfolded proteins, some of these improperly folded proteins
can produce toxic masses.
In the case quality control mechanism by aaRSs, three different
environmental conditions had been selected:
1. Low ratio of cognate amino acids to non-cognate amino
acids due to the short supply of nutrients and probably
due to other stresses
2. Induction of misfolded protein and heat shock protein response
3. The Stress conditions that result in a decrease in the rate
of cell growth, ultimately lessen the appropriate dilution
of aggregated and misfolded protein during the growth of
cells and permit the buildup of toxic aggregates.
Complications with the Non-Cognate Amino
Acids
Quality control mechanism by aaRSs appears to sustain the viability
of cells even when there are significant levels of non-cognate
amino acids that can enhance tRNA mischarging. aaRSs
quality control is required by all the three domains of life when
there is an excess of non-cognate and nonstandard amino acids.
For instance, mutant AlaRS in the mouse fibroblast cells
is-acylates tRNA Ala with amino acid Serine, but the lack of editing
pocket for Ser-tRNA. The Sulfolobus solfataricus (Archean)
is responsible for the misincorporation of Serine [52,54].
Besides this, in the E. coli cells, mutations in valRS or editing
activity of leuRS results in the loss of viability in the occurrence
of surplus α-aminobutyrate (nonstandard amino acid) or the
non-cognate amino acid Ile. However, the limited knowledge
about the occurrence of perturbed amino acids may limit the
extraction of possible conclusions from this data [11].
Experiments also propose that the cell response towards altered
amino acid ratios may vary when the cells are provided
with amino acids either exogenously or manufactured endogenously.
For example, the ratio of endogenously synthesized
Phe to Tyr decreases up to fourfold due to transferring of yeast
from a complex growth medium to minimal growth medium,
which results in an increased requirement for PheRS quality
control for attaining the optimal growth of cells (N.m.R. and
mi., unpublished observations). The pool of amino acids produced
endogenously can be affected by the cell stresses like
variations in the carbon sources but their consequences on the
accuracy of translation are unidentified [2].
Upregulation of Misfolded Proteins Synthesis
Misfolding or unfolding of proteins is crucially associated
with environmental conditions. Environmental conditions
for instance heat cause the unfolding of protein which effectively
harms the growth of cells, and it causes defects in the
translational quality control of the cell. The defects caused by
mistranslation in the translational quality control mechanism
promote the unfolding of proteins. It is found that unfolded
protein response, also acknowledged as the heat shock response,
is triggered only in the cells that were expressing aaRSs
with an editing defect [53]. Moreover, the cells expressing
is-acylated aaRSs or aaRSs with an editing defect considerably
increase the formation of heat shock proteins as observed in E. coli strains [53]. Purkinje cells with mutated AlaRS, which
miscalculates tRNAAla with Ser, showed a significant increase
in ubiquitylation and buildup of proteins and boosted production
of heat shock protein [10]. The cells of S. cerevisiae were
purposely engineered to mistranslate leu amino acid as Ser,
this mistranslation resulted from the upregulation of the heat
shock proteins synthesis which helped the engineered cells
to survive at a lethal temperature of 50°c for a longer period
than the wild-type S. cerevisiae cells [55]. Nevertheless, lack
of translational quality control may cause mistranslation to
occur which results enhance the cell’s capability to survive at
lethal temperatures, but it can also cause many growth defects
at sub-lethal temperatures because of high levels of misfolded
proteins in the cell. The cells of E. coli with an editing defect in
IleRS exhibit limited growth of cells at high, but not the lethal
temperatures [55].
Effect of Lacking Quality Control Mechanisms
on Growth Conditions
Lack of quality control mechanisms by aaRSs may cause some
effects in the cells; some of these effects cannot be instantly
described that either these are occurring due to surplus
non-cognate amino acids or misfolded proteins. However, slow
growth is found to be an important aspect that seems to unite
these effects. The growth of cells is decelerated by unfavorable
environmental conditions, limited supply of cell essential nutrients,
and accumulation of toxic masses. The problems exiting
in controlling mechanisms can be the leading cause of slow
growth in cells by making them more vulnerable to environmental
stresses. Such as, E. coli strains to have IleRS with an
editing defect were reported as more vulnerable to those antibiotics
that can obstruct their ribosomes activity, formation of the cell wall, and DNA replication [55]. Moreover, when these
similar cells having IleRS with editing defects were placed on a
standard growth medium for several days, a great number of
cells with this defect became rifampicin-resistant [56].
Aging bacterial colonies arose with many mutations that cause E. coli cells to develop resistance to rifampicin because these
mutations bring favorable changes that support the growth
of cells during slow growth situations [57]; this recommends
that cells with mutated aaRSs show more severe defects in cell
growth than the wild type cells present in the environment. But
the significance of aaRSs quality control mechanisms is unidentified
under the slow cell growth conditions. Though hair loss
and neurodegeneration were observed in mice having mutated
AlaRS editing site [10], these side effects may be a result of a
buildup of accumulated and ubiquitylated proteins present in
the cell, however, this mutation in the AlaRS editing site also
results in stimulating the production of unfolded proteins in
the Purkinje cells. As if all cells are able of inheriting mutations
and therefore, they become susceptible to consequences of
misfolded proteins buildup, but the degenerative phenotype
is only shown by the Purkinje cells. This effect suggests that
some cell-specified physiological variations are required for the
translational quality control mechanisms in the cell. In terminally
differentiated Purkinje cells, cell division cannot dilute the
unfolded protein response which may cause their more rapid
accumulation in the cell.
Conclusion
As now we thoroughly understand (at the molecular level) that
how the quality control mechanism of mistranslation allows us
to understand better that how simple changes in physiological
conditions determine mistranslation levels. Such variation in
oxidative stress level and also in amino acid pool affects levels
of tRNA mischarging and effectiveness of proofreading. In
the future, we are needed to develop more and more modified
techniques for measuring the rates of mistranslation regularly
or to understand this phenomenon precisely that how translational
accuracy is affected by external and internal factors.
It appears that living beings have advanced a translational fidelity
that is finely adjusted and fluctuates as per that specific
living being’s natural niche. Genetic code variations impact the
cell at the most basic, informational level. These variations provide
important regulatory mechanisms, allow cells to increase
genetic diversity, and reveal novel tools for synthetic biology
applications. Another exciting area for future study can be the
tRNA variations’ effect on human biology. It is now evident that
mistranslation plays a vital role in both normal development
and the pathogenesis of the human disease, thus emphasizing
the potential of therapeutics targeting this pathway.
Acknowledgment
None
Conflict of Interest
None
REFERENCES
- Hoffman KS, O'Donoghue P, Brandl CJ (2017) Mistranslation: From adaptations to applications. Biochim Biophys Acta Gen Subj 1861(11):3070-3080.
[Crossref] [Google Scholar] [PubMed]
- Reynolds NM, Lazazzera BA, Ibba M (2010) Cellular mechanisms that control mistranslation. Nat Rev Microbiol 8(12):849-856.
[Crossref] [Google Scholar] [PubMed]
- Crick F (1970) Central dogma of molecular biology. Nature 227(5258):561-563.
[Crossref] [Google Scholar] [PubMed]
- Moghal A, Mohler K, Ibba M (2014) Mistranslation of the genetic code. FEBS Lett 588(23):4305-4310.
[Crossref] [Google Scholar] [PubMed]
- Markiewicz P, Kleina LG, Cruz C, Ehret S, Miller JH (1994) Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as "spacers" which do not require a specific sequence. J Mol Biol 240(5):421-433.
[Crossref] [Google Scholar] [PubMed]
- Guo HH, Choe J, Loeb LA (2004) Protein tolerance to random amino acid change. Proc Natl Acad Sci U S A 101(25):9205-9210.
[Crossref] [Google Scholar] [PubMed]
- Ibba M, Söll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69:617-650.
[Crossref] [Google Scholar] [PubMed]
- Schimmel P (2011) Mistranslation and its control by tRNA synthetases. Philos Trans R Soc Lond B Biol Sci 366(1580):2965-2971.
[Crossref] [Google Scholar] [PubMed]
- de Pouplana LR, Santos MA, Zhu JH, Farabaugh PJ, Javid B (2014) Protein mistranslation: Friend or foe? Trends Biochem Sci 39(8):355-362.
[Crossref] [Google Scholar] [PubMed]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, et al. (2006) Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443(7107):50-55.
[Crossref] [Google Scholar] [PubMed]
- Nangle LA, Motta CM, Schimmel P (2006) Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol 13(10):1091-1100.
[Crossref] [Google Scholar] [PubMed]
- Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ (2008) Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135(4):679-690.
[Crossref] [Google Scholar] [PubMed]
- Drummond DA, Wilke CO (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134(2):341-352.
[Crossref] [Google Scholar] [PubMed]
- Novoa EM, Pavon-Eternod M, Pan T, de Pouplana LR (2012) A role for tRNA modifications in genome structure and codon usage. Cell 149:202-213.
[Crossref] [Google Scholar] [PubMed]
- Bratulic S, Toll-Riera M, Wagner A (2017) Mistranslation can enhance fitness through purging of deleterious mutations. Nature Commun 8:15410.
[Crossref] [Google Scholar] [PubMed]
- Gout JF, Thomas WK, Smith Z, Okamoto K, Lynch M (2013) Large-scale detection of in vivo transcription errors. Proc Natl Acad Sci U S A 110:18584-9.
[Crossref] [Google Scholar] [PubMed]
- Traverse CC, Ochman H (2016) Conserved rates and patterns of transcription errors across bacterial growth states and lifestyles. Proc Natl Acad Sci U S A 113:3311-6.
[Crossref] [Google Scholar] [PubMed]
- Swanson R, Hoben P, Sumner Smith M, Uemura H, Watson L, et al. (1988) Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science 242:1548-51.
[Crossref] [Google Scholar] [PubMed]
- Kramer EB, Farabaugh PJ (2007) The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13:87-96.
[Crossref] [Google Scholar] [PubMed]
- Drummond DA, Wilke CO (2009) The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 10:715-24.
[Crossref] [Google Scholar] [PubMed]
- Whitehead DJ, Wilke CO, Vernazobres D, Bornberg-Bauer E (2008) The look-ahead effect of phenotypic mutations. Biol Direct 3:18.
[Crossref] [Google Scholar] [PubMed]
- Bratulic S, Gerber F, Wagner A (2015) Mistranslation drives the evolution of robustness in TEM-1 beta-lactamase. Proc Natl Acad Sci U S A 112:12758-63.
[Crossref] [Google Scholar] [PubMed]
- Griswold CK, Masel J (2009) Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex. PLoS Genet 5:e1000517.
[Crossref] [Google Scholar] [PubMed]
- Yanagida H, Gispan A, Kadouri N, Rozen S, Sharon M, et al. (2015) The evolutionary potential of phenotypic mutations. PLoS Genet 11:e1005445.
[Crossref] [Google Scholar] [PubMed]
- Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC (2001) Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol 8:238-42.
[Crossref] [Google Scholar] [PubMed]
- Manickam N, Nag N, Abbasi A, Patel K, Farabaugh PJ (2014) Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA 20:9-15.
[Crossref] [Google Scholar] [PubMed]
- Fan Y, Wu J, Ung MH, De Lay N, Cheng C, et al. (2015) Protein mistranslation protects bacteria against oxidative stress. Nucleic Acids Res 43:1740-1748.
[Crossref] [Google Scholar] [PubMed]
- Ballesteros M, Fredriksson A, Henriksson J, Nystrom T (2001) Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. Embo j 20:5280-9.
[Crossref] [Google Scholar] [PubMed]
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, et al. (2009) Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462:522-6.
[Crossref] [Google Scholar] [PubMed]
- Ling J, Soll D (2010) Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci U S A 107:4028-33.
[Crossref] [Google Scholar] [PubMed]
- Dunlop RA, Cox PA, Banack SA, Rodgers KJ (2013) The non-protein amino acid BMAA is misincorporated into human proteins in place of L-serine causing protein misfolding and aggregation. PLoS One 8:e75376.
[Crossref] [Google Scholar] [PubMed]
- Pablo J, Banack SA, Cox PA, Johnson TE, Papapetropoulos S, et al. (2009) Cyanobacterial neurotoxin BMAA in ALS and Alzheimer’s disease. Acta Neurol Scand 120:216-225.
[Crossref] [Google Scholar] [PubMed]
- Ling J, Reynolds N, Ibba M (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol 63:61-78.
[Crossref] [Google Scholar] [PubMed]
- Martinis SA, Boniecki MT (2010) The balance between pre‐and post‐transfer editing in tRNA synthetases. FEBS Lett 584:455-459.
[Crossref] [Google Scholar] [PubMed]
- Schmidt E, Schimmel P (1994) Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science 264:265-266.
[Crossref] [Google Scholar] [PubMed]
- Minajigi A, Deng B, Francklyn CS (2011) Fidelity escape by the unnatural amino acid β-hydroxynorvaline: An efficient substrate for Escherichia coli threonyl-tRNA synthetase with toxic effects on growth. Biochemistry 50:1101-1109.
[Crossref] [Google Scholar] [PubMed]
- Dulic M, Cvetesic N, Perona JJ, Gruic-Sovulj I (2010) Partitioning of tRNA-dependent editing between pre-and post-transfer pathways in class I aminoacyl-tRNA synthetases. J Biol Chem 285:23799-23809.
[Crossref] [Google Scholar] [PubMed]
- Wydau S, Ferri-Fioni ML, Blanquet S, Plateau P (2007) GEK1, a gene product of Arabidopsis thaliana involved in ethanol tolerance, is ad-aminoacyl-tRNA deacylase. Nucleic Acids Res 35:930-938.
[Crossref] [Google Scholar] [PubMed]
- Ling J, So BR, Yadavalli SS, Roy H, Shoji S, et al. (2009) Resampling and editing of mischarged tRNA prior to translation elongation. Mol cell 33:654-660.
[Crossref] [Google Scholar] [PubMed]
- Guo M, Chong YE, Shapiro R, Beebe K, Yang XL, et al. (2009) Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature 462:808-812.
[Crossref] [Google Scholar] [PubMed]
- Wydau S, van der Rest G, Aubard C, Plateau P, Blanquet S (2009) Widespread distribution of cell defense against D-aminoacyl-tRNAs. J Biol Chem 284:14096-14104.
[Crossref] [Google Scholar] [PubMed]
- Dale T, Uhlenbeck OC (2005) Amino acid specificity in translation. Trends Biochem Sci 30:659-665.
[Crossref] [Google Scholar] [PubMed]
- Kaminska M, Havrylenko S, Decottignies P, Le Maréchal P, Negrutskii B, et al. (2009) Dynamic organization of aminoacyl-tRNA synthetase complexes in the cytoplasm of human cells. J Biol Chem 284:13746-13754.
[Crossref] [Google Scholar] [PubMed]
- Kyriacou SV, Deutscher MP (2008) An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol Cell 29:419-427.
[Crossref] [Google Scholar] [PubMed]
- Ogle JM, Ramakrishnan V (2005) Structural insights into translational fidelity. Annu Rev Biochem 74:129-177.
[Crossref] [Google Scholar] [PubMed]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H (2010) Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 466:329-333.
[Crossref] [Google Scholar] [PubMed]
- Ling J, Yadavalli SS, Ibba M (2007) Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA 13:1881-1886.
[Crossref] [Google Scholar] [PubMed]
- Effraim PR, Wang J, Englander MT, Avins J, Leyh TS, et al. (2009) Natural amino acids do not require their native tRNAs for efficient selection by the ribosome. Nat Chem Biol 5:947-953.
[Crossref] [Google Scholar] [PubMed]
- Sørensen MA, Elf J, Bouakaz E, Tenson T, Sanyal S, et al. (2005) Over expression of a tRNA Leu isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J Mol Biol 354:16-24.
[Crossref] [Google Scholar] [PubMed]
- Dix DB, Thompson RC (1989) Codon choice and gene expression: Synonymous codons differ in translational accuracy. Proc Natl Acad Sci U S A 86:6888-6892.
[Crossref] [Google Scholar] [PubMed]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, et al. (2009) The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326:688-694.
[Crossref] [Google Scholar] [PubMed]
- Ling J, Roy H, Ibba M (2007) Mechanism of tRNA-dependent editing in translational quality control. Proc Natl Acad Sci U S A 104:72-77.
[Crossref] [Google Scholar] [PubMed]
- Ruan B, Palioura S, Sabina J, Marvin-Guy L, Kochhar S, et al. (2008) Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci U S A 105:16502-16507.
[Crossref] [Google Scholar] [PubMed]
- Korencic D, Ahel I, Schelert J, Sacher M, Ruan B, et al. (2004) A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc Natl Acad Sci U S A 101:10260-10265.
[Crossref] [Google Scholar] [PubMed]
- Bacher JM, de Crécy-Lagard V, Schimmel PR (2005) Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc Natl Acad Sci U S A 102:1697-1701.
[Crossref] [Google Scholar] [PubMed]
- Bacher JM, Schimmel P (2007) An editing-defective aminoacyl-tRNA synthetase is mutagenic in aging bacteria via the SOS response. Proc Natl Acad Sci U S A 104:1907-1912.
[Crossref] [Google Scholar] [PubMed]
- Wrande M, Roth JR, Hughes D (2008) Accumulation of mutants in “aging” bacterial colonies is due to growth under selection, not stress-induced mutagenesis. Proc Natl Acad Sci U S A 105:11863-11868.
[Crossref] [Google Scholar] [PubMed]
Citation: Shad M, Malik A, Javid M, Din S, Shakoor S, et al. (2022) Mistranslation of Genetic Codon and Their Controlling Mechanism in Cell. Biochem Mol Bio J. 08:51.
Copyright: Shad M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.





