Research Article - (2024) Volume 10, Issue 5
Received: 09-Apr-2020, Manuscript No. IPCE-24-3748; Editor assigned: 14-Apr-2020, Pre QC No. IPCE-24-3748 (PQ); Reviewed: 28-Apr-2020, QC No. IPCE-24-3748; Revised: 21-Jun-2024, Manuscript No. IPCE-24-3748 (R); Published: 19-Jul-2024, DOI: 10.21767/2472-1158-24.10.41
Epigenetic mutations of some peculiar genic functions play a crucial role in gliomagenesis. One of the most important in neurooncological pharmaco-epigenetics is represented by the MGMT promoter methylation.
We evaluated the MGMT gene promoter methylation status by real-time PCR and sequencing of the entire promoter. The samples from 25 patients, men and women with an average age of 55 years, with intracranial tumors, were consecutively evaluated. By subsequent and definitive histological typing, the patient distribution showed 11 patients with GBM, 3 with HGG, 3 with LGG and 8 with brain metastases.
Analyzing the MGMT gene promoter methylation status of a non-homogeneous series of 25 consecutive patients, 5 patients had High-Grade Glioma (HGG) with unmethylated MGMT gene promoter and 4 patients had glioblastoma with almost complete MGMT gene promoter methylation, higher than the cut-off value of 80%, indicated as a possible threshold for highly positive response to temozolomide. The remaining patients (including those with metastatic disease) presented partial methylation, which does not indicate, at the current moment, any peculiar prognostic and therapeutic indications.
The present study has confirmed the validity of this method, both in terms of sensitivity and specificity, for the evaluation of MGMT gene promoter methylation status, based on the amplification of specific DNA methylation and not on real-time PCR and sequencing of the en-tire MGMT gene promoter sequence. These data certainly open a new scenario up in oncology, which now combines with the study of the tumor epigenomics.
Glioblastoma; Methylation; Promoter; Temozolomide; Epigenomics
A crucial role in gliomagenesis high genetic and molecular variability of High-Grade Gliomas (HGG) and Glioblastomas (GBM) in particular and in response to antineoplastic drugs has been played by epigenetic mutations of some peculiar genic functions. One of the most important in neurooncologic pharmacoepigenetics is represented by the 06- Methylguanine-DNA-Methyltransferase (MGMT) promoter methylation [1]. Many studies have reported that, among patients affected by GBM, those with MGMT promoter methylation have a higher survival and a better response to chemotherapy with alkylating agents. The aim of the study was to focus on the epigenetic, translational role and importance of 06-Methylguanine-DNA-Methyltransferase (MGMT) methylation status into gliomagenesis as a predictive factor for the therapeutic response and efficacy of chemotherapy with alkylating agents, such as temozolomide. This objective was achieved through the evaluation of the MGMT gene promoter methylation status by real-time PCR and sequencing of the entire promoter [2].
DNA Extraction
The MGMT gene promoter methylation status have been analyzed on a fresh sample, isolated from the surgically removed lesion after extemporaneous histological examination, biopsy specimens of tumor tissue, removed during brain surgery (removal of suspected high-grade glial tumor or glial tumor already typed by extemporaneous histopathological study), undergo as fresh samples the study in question. This protocol is part of the current treatment guidelines for high-grade gliomas.
The manual method used for DNA extraction provides for the use of TRIzol® reagent. TRI-zol® reagent isolates RNA, DNA and proteins in a fast and simultaneous way. It can be used with large or small amounts of tissue or cells and multiple samples can be extracted simultaneously. TRIzol® is a monophasic solution of phenol and guanidine isothiocyanate in acid environment that preserves the integrity of RNA, the most perishable nucleic acid and at the same time separates the protoplasm into its main components. Normally, to each 50 mg-100 mg sample, 1 ml of TRIzol® is added. The whole is then transferred in an eppendorf test tube, where 200 μl of chloroform-a powerful deproteinizing compound are added, to remove any molecules that may contaminate the DNA, to prevent them from interfering with the sub-sequent treatments.
After 15 minutes of incubation at room temperature, the samples are centrifuged at 15000 rpm for 15 minutes [3].
The centrifugation separates the mixture into 3 phases: 1) Proteins: (Organic phase) red lower phase; 2) DNA: (Interphase) white central phase; 3) RNA: (Aqueous phase) transparent upper phase. The supernatant containing RNA is collected and transferred to a new eppendorf tube, precipitated in isopropanol and stored at -80°C. In the eppendorf tube containing proteins and DNA, 300 μl of ethanol absolute are added, to precipitate the DNA. The samples are agitated with a vortex mixer and let to incubate at room temperature for 10 minutes. Afterwards, they are centrifuged at 2000 xg for 5 minutes and after removal of the supernatant, 1 ml of a solution containing sodium citrate and ethanol 10% is added.
This saline solution interacts with the DNA’s electrical charge, allowing its precipitation in alcohol. The samples are then incubated in ice for 30 minutes and then centrifuged for 5 minutes at 2000 xg. This step is performed a second time, then the supernatant is removed and 1 ml of ethanol 75% is added, to remove salts from the pellet that resulted. After 10 minutes of incubation at room temperature, the samples are centrifuged for 5 minutes at 2000 xg and after removal of the supernatant, the pellet is dissolved in 61 μl of 8 mM NaOH. This is an alkaline solution that ensures the complete dissolution of the DNA pellet. To remove insoluble material, the mixture is centrifuged at 12000 xg for 10 minutes. The supernatant containing DNA is finally transferred to a new eppendorf test tube [4].
DNA Quantitation
The extracted DNA is quantitated with the spectrophotometer (Nanodrop), which allows to identify and quantitate a substance by analyzing its light absorption spectrum. With this tool we performed the readings of our DNA samples, to know their concentrations. This technique takes advantage of the peculiar absorption spectrum of the UV radiation created by the cyclic chains of the nitrogenous bases.
Treatment of DNA with Sodium Bisul ite
The DNA, extracted and quantitated spectrophotometrically, is treated using the epitect bisulfite® kit (Qiagen). This kit uses an advanced procedure based on a new desulfonation method to efficiently convert unmethylated Cytosine (C) in Uracil (U), whereas methylated cytosine is protected from conversion.
After Polymerase Chain Reaction (PCR), the DNA sequence treated with sodium bisulfite contains C residues only if it were methylated; all other C is converted into Thymine (T). Thus, a sample of unmethylated DNA will not contain any C residues after treatment. A comparison between a DNA sequence treated with sodium bisulfite and an untreated DNA sequence allows a more precise identification of all the methylated C residues [5].
The DNA, converted and purified, can be used later for sequencing, cloning, digestion with restriction enzymes etc. With this kit, we must dilute 200 mg of genomic DNA (gDNA) in deionized water to a final volume of 45 μl. We then add 5 μl of denaturation buffer to denaturate the DNA. The volume is now 50 μl. The samples are incubated at 37°C for 15 minutes. At this point the reagent, provided in powder form, is prepared for the conversion (conversion reagent). To the test tube containing the powder, 750 μl of deionized water and 210 μl of denaturation buffer are added. To thoroughly dissolve the powder, the tube is put on a vortex mixer for 1 minute and then rested for 2 minutes. This procedure is repeated 5 times. When the powder is dissolved, we add 100 μl of this reagent to the DNA samples, which are now denatured, because of the conversion of unmethylated cytosine to uracil [6]. The volume is now 150 μl. The DNA samples must then incubate in the dark at 50°C for 12 hours. After the incubation period, during which the unmethylated C is converted into an intermediate sulfonate, the DNA must be purified. Purification columns are inserted into Eppendorf test tubes (supplied with the kit). In each column we put 200 μl of deionized water and 150 μl of sample. The samples are centrifuged at 500 x g for 20 minutes and then the supernatant is discarded. Afterwards, 350 μl of deionized water is added to the upper chamber of the column and the samples are centrifuged at 500 x g for 20 minutes. The supernatant is discarded again. These last three steps are repeated a second time. 350 μl of 0.1 M NaOH are added and left to incubate for 5 minutes. During this incubation step, the intermediate sulfonate is converted into uracil. The mixture is centrifuged for 20 minutes at 500 x g and the supernatant is discarded. Subsequently, 350 μl of deionized water are added and we centrifuge at 500 x g for 20 minutes. Then 50 μl of TE buffer (Tris EDTA) are added, the solution is mixed with a pipette and left to incubate for 5 minutes. After the incubation period has expired, the columns must be inverted and inserted into new eppendorf test tubes. Finally, the mixture is centrifuged at 1000 x g for 1 minute and genomic DNA treated with bisulfite, ready for PCR, can be found on the bottom of the test tube. If the samples are not used immediately, they should be stored at 4°C and can be kept and used up to a year [7].
DNA Amplification
The classic PCR is a qualitative assay that allows an assessment of the products only at the end of the experiment (end-point measurement), that is when, due to the plateau effect, the final quantity of product is not correlated anymore with the starting amount.
In real-time PCR, instead, the measurement is carried out during the exponential phase, during which the influence of the reaction variables is minimal and the number of cycles required to obtain a detectable fluorescence signal significantly greater than the background noise is correlated to the starting amount of template DNA In the specific case, the product (DNA) amplified by endpoint PCR, after treatment with sodium bisulfite, is also sequenced (this allows the study of the entire promoter of the MGMT gene) and can be subjected to a further amplification with real-time PCR, using two different pairs of primers, specific respectively for methylated and unmethylated DNA [8].
Evaluation of MGMT Gene Promoter Methylation Status
The evaluation of MGMT gene promoter methylation status was carried out by means of a first PCR amplification of the DNA treated with bisulfite. At this point, the amplified product is subjected to electrophoresis on agarose gel and subsequently sequenced (Figure 1).
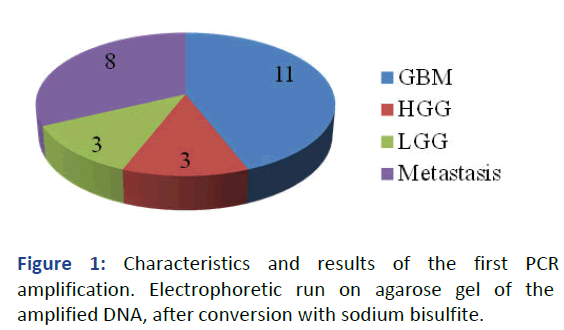
Figure 1: Characteristics and results of the first PCR amplification. Electrophoretic run on agarose gel of the amplified DNA, after conversion with sodium bisulfite.
On the amplified product was then performed a second realtime PCR with two pairs of primers, respectively for methylated and unmethylated DNA (Figure 2) [9].
Figure 2: Second DNA amplification with real-time PCR.
Sequencing by Capillary Electrophoresis
The classic sanger sequencing (di-deoxy sequencing) which exploits the ability of DNA polymerase to incorporate 2', 3' dideoxy-nucleotides (ddNTPs) into the single-stranded nascent DNA instead of the normal Nucleotide Triphosphates (dNTPs), with the consequent interruption of the polymerization reaction has now mostly given way to new sequencing techniques that include the use of fluorescent markers, which automate the reading process of the amplified products after capillary electrophoresis. These methods use two different chemical tools: Labeled primers and marked ddNTPs (or terminators), using in both cases four different fluorochromes, which can be excited with electromagnetic radiation of same or similar wavelength, but with peaks in the emission spectra sufficiently distinct that they can be discriminated by the sequencer’s detection system. On the other hand, with labeled primers, four independent PCR reactions are started each with limited quantities of a single ddNTP in which the primer will be labeled on the 5' with a single fluorochrome, whose signal will then be matched to that particular dideoxynucleotide. The products, after column based purification, are subjected to capillary electrophoresis within a sequencer equipped with a laser. As the fragments intercept the laser light, a signal is detected by the fluorescence of the fluorophore bound to the primer, which shows that an eluted fragment at that particular time and of that determined length ends with that particular base [10].
The fluorescence signals detected continuously and represented by the software as a function of elution time take the name of electropherograms. As for labeled terminators, a single PCR is started, in which the various ddNTPs, each marked as usual, although in a mixture, will confer to each fragment that will emit a characteristic fluorescence signal. In both cases the sequence revealed will be complementary to the DNA strand in the 5’-3’ direction (Figure 3).
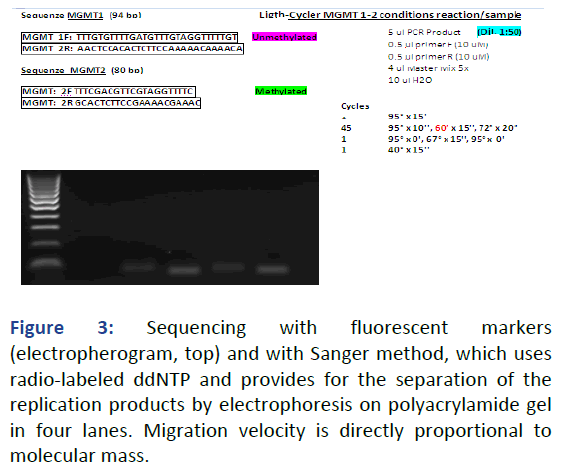
Figure 3: Sequencing with fluorescent markers (electropherogram, top) and with Sanger method, which uses radio-labeled ddNTP and provides for the separation of the replication products by electrophoresis on polyacrylamide gel in four lanes. Migration velocity is directly proportional to molecular mass.
For this project work, we have analyzed the MGMT promoter methylation status of representative samples of fresh tumor tissue taken after neurosurgical procedures of resection. The samples from 25 patients, men and women with an average age of 55 years, with intracranial tumors, were evaluated consecutively and after extemporaneous intraoperative histological examination. By subsequent and definitive histological typing, the patient distribution showed 11 patients with GBM, 3 with HGG, 3 with LGG and 8 with secondary brain tumor (brain metastases) (Figure 4).
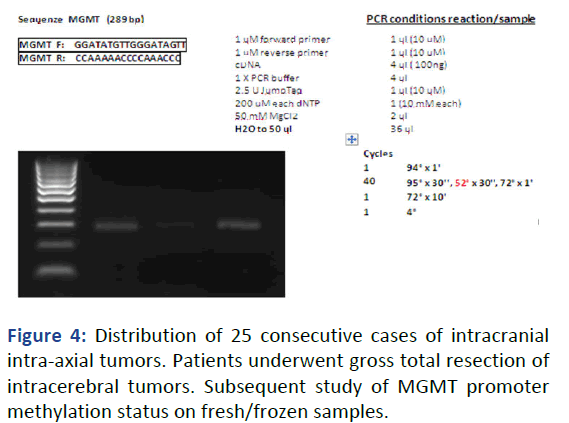
Figure 4: Distribution of 25 consecutive cases of intracranial intra-axial tumors. Patients underwent gross total resection of intracerebral tumors. Subsequent study of MGMT promoter methylation status on fresh/frozen samples.
Analyzing the MGMT gene promoter methylation status of a non-homogeneous series of 25 consecutive patients, 5 patients had High-Grade Glioma (HGG+GBM) with unmethylated MGMT gene promoter thus they were not eligible candidates for chemotherapy with temozolomide or other alkylating agents and 4 patients had GBM with almost complete MGMT gene promoter methylation, higher than the cutoff value of 80%, indicated as a possible threshold for highly positive response to temozolomide. The remaining patients (including those with metastatic disease) presented partial methylation, which does not indicate, at the current moment, any peculiar prognostic and therapeutic indications. From what has previously emerged, it follows that it is important to correctly divide patients into subgroups characterized by their neoplastic disease’s genomic and epigenomic features, in order to gain significantly predictive data, which will be of help during therapy. The data from our study should be confirmed by expanding the series and possibly by considering other genetic factors involved in the response of glioblastoma multiforme to treatment with alkylating agents [11].
GBM is a malignant tumor that originates from glial cells, probably from a cluster of pathological neuroglial stem cells and the most common and most aggressive of all brain tumors, as it has distinct characteristics and is highly recurrent and chemoradioresistant. For these reasons, the average survival of patients with glioblastoma multiforme varies around 12-14 months. In recent years, the clinical use of chemotherapy with alkylating agents has improved the overall prognosis, although very slightly. Alkylating agents cause DNA damage by determining the pairing of thymine instead of cytosine, resulting in the apoptosis of the cancer cell. The clinical usage of alkylating agents is limited by the activity of the DNA repair system, by enzymes with capacity to dealkylate DNA. Among these proteins MGMT is one of the most important, as it protects the tumor cells by subtracting A statistical analysis showed a significant difference in progression free survival rates between the group of patients with survival lower than 15 months and those with survival higher than 25 months. A second statistical analysis, which aimed to establish a methylation cutoff, above which the survival of patients appeared to be the highest, has shown a statistically significant difference in survival between the group of patients with methylation values higher than 80% and the other two groups. Therefore, this analysis revealed that there is a potential MGMT promoter methylation cutoff at 80%, above which the survival of patients enrolled in our study was found to be higher than 25 months [12].
Our dual study protocol, sequencing of the entire gene promoter and real-time PCR amplification of methylated and unmethylated DNA, represents an important advance in the study of MGMT promoter methylation status, significantly increasing both the sensitivity and specificity of this key epigenetic marker study. These improvements provide a very important predictive indication of the response to treatment with alkylating agents, with temozolomide for high grade gliomas [13].
The present study has confirmed the validity of this method, both in terms of sensitivity and specificity, for the evaluation of MGMT gene promoter methylation status, based on the amplification of specific DNA methylation and not on realtime PCR and sequencing of the en-tire MGMT gene promoter sequence. This may be a good foundation for the development of this method in future clinical applications, as it provides important parameters for the evaluation of clinical responses to treatment with alkylating agents and at the same time avoids the prescription of this therapy to patients with low or no MGMT gene promoter methylation also in consideration of the very high possibility of toxic effects opting in those instances for other therapeutic protocols.
Future studies will be aimed at the search of any other factors related to a better response to alkylating agents, trying in this way to improve the survival to chemotherapy of patients with glioblastoma multiforme. These data certainly open a new scenario up in oncology, which now combines with the study of the tumor epigenomics.
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Raudino G, Umana G, Raudino F, Lacognata V, Iemmolo R, et al. (2024) Molecular Bases of Gliomagenesis and Gene Sequencing-Based Study of the 06-Methylguanine-DNA-Methyltransferase Promoter Methylation: Prognostic and Therapeutic Implications. J Clin Epigen. 10:41.
Copyright: © 2024 Raudino G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.