Short Communication - (2016) Volume 1, Issue 1
Gurudeeban Selvaraj*, Satyavani Kaliamurthi and Ramanathan Thiruganasambandam
Centre of Advanced Study in Marine Biology, Faculty of Marine Sciences, Annamalai University, Parangipettai 608502, Tamil Nadu, India
Corresponding Author:
Dr. S. Gurudeeban
Centre of Advanced Study in Marine Biology, Faculty of Marine Sciences, Annamalai University, Parangipettai 608502, Tamil Nadu, India
Tel: 91-978-9445377
E-mail: gurudeeb99@gmail.com
Received: January 30, 2016; Accepted: March 04, 2016; Submitted: March 07, 2016
Citation: Selvaraj G, Kaliamurthi S, Thiruganasambandam R. Molecular Docking Studies of Rutin on Matrix Metalloproteinase. Insights Biomed. 2016, 1:1.
Matrix metalloproteinase (MMP) also known as matrixins, play an important role in wound healing, tissue engineering and embryogenesis. The action of MMP inhibited by several endogenous tissue inhibitors but the pathological condition requires exogenous inhibitor from natural or synthetic source. Therefore, the present study is aimed to evaluate the inhibitory effect of rutin on metalloproteinase. The active sites of target proteins were predicted through 3DLignadsite. Based on the Lamarckian genetic algorithm, the energy minimized rutin was docked with MMP2, MMP3, MMP8 and MMP12 using Autodock 4.2. The result indicates the interaction of rutin exhibited binding free energy of MMP2 as -10.19kcalmol-1 on GLU, MMP8 as -8.83kcalmol-1 on LEU, THR, MMP3 as -8.83kcalmol-1 on ALA, HIS and MMP12 as -6.08kcalmol-1 CYS and HIS57 residues. We conclude rutin as an effective matrix metalloproteinase inhibitor. Further studies will confirm the usage of rutin as a matrix metalloproteinase inhibitor.
Keywords
Active site residues; Elastase; Molecular docking; Rutin; Wound healing
Introduction.
MMPs are proteolytic and matrix degrading enzymes play multiactions in the pathology of the nervous system. Among the various factors, the extracellular matrix (ECM) made from fibrin, elastin, hyaluronan, type I and type III collagen are combined together to form a fibrenetwork to maintain the structural integrity [1]. Matrix metalloproteinase level was increased in diabetic patients who simultaneously causes inflammation, atherogenic effect and increased the tumour necrosis α-factor [2]. In diabetic foot ulcer, the inflammatory response accelerated the synthesis of matrix metalloproteinase to degrade the ECM fibrous structural proteins, which decreased the blood supply, delayed wound healing, atherosclerosis, and gangrene of the foot finally leading to amputations [3]. MMP contained different class of enzymes, especially gelatinase (MMP2), stromelysins (MMP3), collagenase (MMP8), and metalloelastase (MMP12) involved in the tissue repairing process [4].
In silico studies will provide a high-quality interaction between the ligand and receptor. This approach is now time-consuming, reasonable; minimize the cost and side effects when entered into clinical studies [5]. Rutin is also known as Quercetin-3-Orutinoside, sophorin and rutoside. Number of studies reported on the biological activities of rutin, includes hypoglycaemic, antidiabetic, osteoblast stimulant, free radical scavenging, antioxidant, anti-depressant, anti-asthmatic and anti-nociceptive effect [6-10]. In another hand rutin loaded solid lipid nanoparticles reported that it improves bioavilability and wound healing capacity in diabetic animal models [11-12]. Quercetin derivatives were reported as a therapeutic agent for the treatment of neutrophildependent inflammatory diseases [13]. Using model chemistry approaches like density functional theory the rutin moment, solubility and polarizability were calculated [14]. The clinical trials of a synthetic MMP inhibitor, Batimastat was not successful because of its adverse effects [15]. The role of plant metabolites on MMP inhibitory action was still increasing because of their lesser side effects. We reported the inhibitory effect of E. agallocha extracts on gelatinase, stromelysins, collagenase, and elastase and rutin was identified as the active constituent in the potential ethanolic extract [16]. With the continuation, the present study aimed to evaluate the molecular interaction of rutin with matrix metalloproteinase using the computational method.
Methods and Methods
Preparation of receptor
Three-dimensional structure of four different matrix metalloproteinase MMP2 (gelatinase), MMP3 (stromelysins), MMP8 (collagenases) and MMP12 (metallo elastase) (PDB ID: 1QIB; 2DIO; 1MNC; 1HNE) was retrieved from the Swiss-Prot database. The structure visualized using the molecular graphics program PyMol® intended for the structural visualization of proteins.
Preparation of ligand
3D structure of rutin was retrieved from the PubChem® database. Optimized ligand was docked into distinguished model using Ligand Fit theory.
Predicting ligand binding-site using 3D ligand site web server
Numerous proteins continue their competence by interacting with drugs or ligands. These include protein cofactors, metabolites and synthetic chemicals. The identification of ligand binding sites is important to determine the protein function on the ligand. 3D ligand site is a web server (https://www.sbg.bio. ic.ac.uk/3dligandsite) was used to identify ligand-binding sites in MMPases.
In silico docking
MGL (Molecular Graphics Laboratory) Tools of AutoDock (4.0) version used to create PDBQT files from traditional PDB files. The receptor file prepared with the addition of polar hydrogens, Kollman charges, and solvation parameters. The precalculated grid maps at the size set at 60, 60, and 60 A° (x, y, and z) to include all the amino acid residues that present in the receptor. The spacing between grid points was 0.575 angstroms. The Lamarckian genetic algorithm (LGA) chosen to confirm maximum of 10 conformers was considered to the docking process with the population size of 150 individuals. The complete docking process performed in Intel CORETM i5, 64 bit Operating System and 4GB RAM in Lenovo Win 7 PC.
Result and Discussion
Active sites
The amino acids (ALA-Alanine; GLY-Glycine; GLN-Glutamine; HISHistidine; SER-Serine; ARG-Arginine; CYS-Cystine; LEU-leucine; THR- Thyronine) in the active sites of MMP2, MMP3, MMP8 and MMP12 were shown in Table 1 and Figure 1.
| Enzyme | Residues | Amino acids |
|---|---|---|
| MMP2 | 159, 161, 163, 181, 184 | GLY, ASP, LEU, ASP, GLU |
| MMP3 | 17, 19, 20, 21, 57, 58, 59, 93, 99, 122, 144 | GLN, LEU, SER, VAL, THR, GLY, ASP, PHE, LEU, PHE, PHE |
| MMP8 | 168, 170, 172, 183, 185, 196 | HIS, ASP, SER, HIS, PHE, HIS |
| MMP12 | 20, 27, 135, 136, 137, 159, 200, 201, 204, 205 | ARG, TRP, GLN, CYS, LEU, ASN, VAL, CYS, ASN, GLY |
Table 1: Active site residues present in the matrix metallo-proteinases.
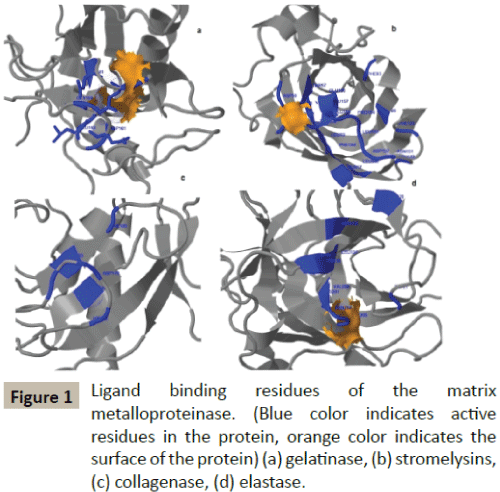
Figure 1: Ligand binding residues of the matrix metalloproteinase. (Blue color indicates active residues in the protein, orange color indicates the surface of the protein) (a) gelatinase, (b) stromelysins, (c) collagenase, (d) elastase.
Molecular Docking
The docking poses were ranked according to their docking scores, list of docked ligands and their corresponding binding poses [17]. After the simulations were complete, the docked structures were analyzed and the interactions were observed. Hydrogen bond interactions and binding distance between the donors and acceptors were measured for the best conformers. Distinct conformation clusters RMSD (Root Mean Square Deviation) -tolerance and Van der Waals scaling factor were found to be 2.0 Å. 1.0 Å respectively.
Rutin interaction with MMP
Docking simulation of rutin ligand into MMP2 produced single cluster of conformers using RMSD-tolerance of 2.0 Å out of 10 docking runs. The ligand showed highest binding energy -10.17 kcal/mol and the reference RMSD value is 65.74. The two hydrogen bond interactions occurred at Glutamine 202 and Glutamine 202 amino acid residues (Table 2 and Figure 2).
| Receptor Protein | No. of H bonds | H-Bond Donor | Length of H-Bond (A) | Binding Energy (kcal/mol) | Inhibition constant Ki (nM) at temperature (298.15 K) | Total Intermolecular Energy (kcal/mol) | Reference RMSD |
|---|---|---|---|---|---|---|---|
| MMP2 | 2 | GLU202 GLU202 |
1.899 2.077 |
-10.19 | 34.14 | -15.00 | 65.74 |
| MMP3 | 2 | LEU70 THR88 |
2.025 2.245 |
-7.79 | 1950 | -13.67 | 88.71 |
| MMP8 | 5 | ALA184 ALA182 HIS222 HIS218 ALA184 |
1.943 2.089 1.918 2.061 2.031 |
-8.83 | 339.66 | -14.32 | 36.17 |
| MMP12 | 2 | CYS58 HIS57 | 2.188 2.001 |
-6.08 | 35120 | -11.53 | 68.15 |
Table 2: Molecular interactions of Rutoside on different target proteins.
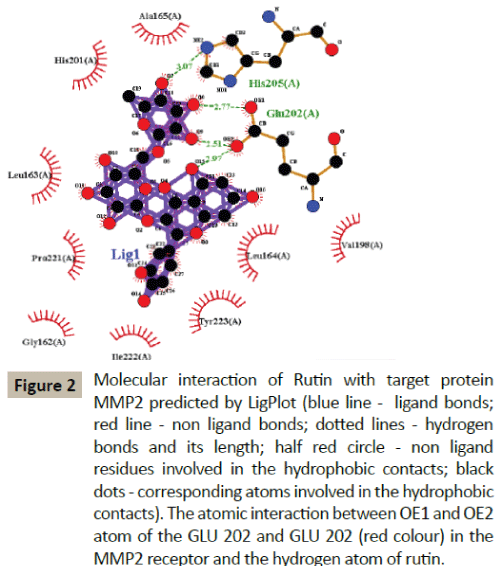
Figure 2: Molecular interaction of Rutin with target protein MMP2 predicted by LigPlot (blue line - ligand bonds; red line - non ligand bonds; dotted lines - hydrogen bonds and its length; half red circle - non ligand residues involved in the hydrophobic contacts; black dots - corresponding atoms involved in the hydrophobic contacts). The atomic interaction between OE1 and OE2 atom of the GLU 202 and GLU 202 (red colour) in the MMP2 receptor and the hydrogen atom of rutin.
Rutin interaction with MMP 3
Docking simulation of rutin ligand into MMP3 produced single cluster of conformers using RMSD-tolerance of 2.0 Å out of 10 docking runs. The ligand showed highest binding energy -7.79 kcal/mol and the reference RMSD value is 88.71. The two hydrogen bond interactions occurred at leucine 70 and Thyronine 88 amino acid residues respectively (Table 2 and Figure 3).
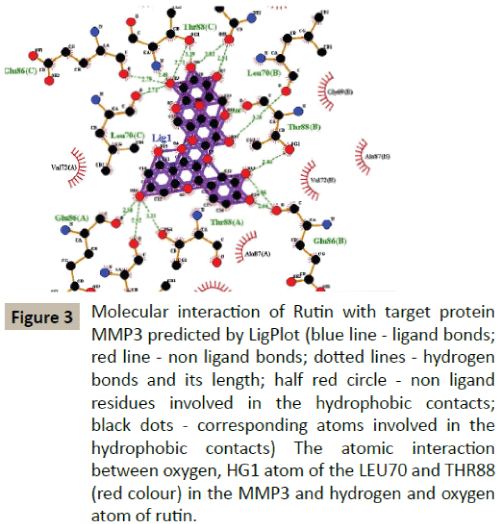
Figure 3: Molecular interaction of Rutin with target protein MMP3 predicted by LigPlot (blue line - ligand bonds; red line - non ligand bonds; dotted lines - hydrogen bonds and its length; half red circle - non ligand
residues involved in the hydrophobic contacts; black dots - corresponding atoms involved in the hydrophobic contacts) The atomic interaction between oxygen, HG1 atom of the LEU70 and THR88 (red colour) in the MMP3 and hydrogen and oxygen atom of rutin.
Rutin interaction with MMP 8
Docking simulation of rutin ligand into MMP8 produced single cluster of conformers using RMSD-tolerance of 2.0 Å out of 10 docking runs. The ligand showed highest binding energy -8.83 kcal/mol and the reference RMSD value is 36.17. The five hydrogen bond interactions occurred at Alanine 184, Alanine 182, Histidine 222, Histidine 218, and Alanine184 amino acid residues respectively (Table 2 and Figure 4).
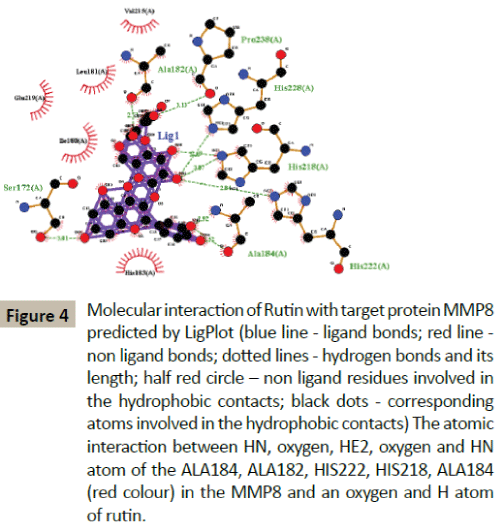
Figure 4: Molecular interaction of Rutin with target protein MMP8 predicted by LigPlot (blue line - ligand bonds; red line - non ligand bonds; dotted lines - hydrogen bonds and its length; half red circle – non ligand residues involved in the hydrophobic contacts; black dots - corresponding atoms involved in the hydrophobic contacts) The atomic interaction between HN, oxygen, HE2, oxygen and HN atom of the ALA184, ALA182, HIS222, HIS218, ALA184 (red colour) in the MMP8 and an oxygen and H atom of rutin.
Rutin interaction with MMP 12
Docking simulation of rutin ligand into MMP12 produced single cluster of conformers using RMSD-tolerance of 2.0 Å out of 10 docking runs. The ligand showed highest binding energy -6.08 kcal/mol and the reference RMSD value is 68.15. The five hydrogen bond interactions occurred at Alanine 184, Alanine 182, Histidine 222, Cystine 58 and Histidine 57 amino acid residues respectively (Table 2 and Figure 5).
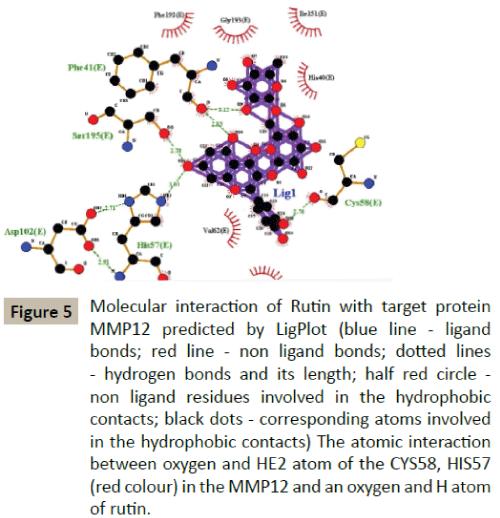
Figure 5: Molecular interaction of Rutin with target protein MMP12 predicted by LigPlot (blue line - ligand bonds; red line - non ligand bonds; dotted lines - hydrogen bonds and its length; half red circle - non ligand residues involved in the hydrophobic contacts; black dots - corresponding atoms involved in the hydrophobic contacts) The atomic interaction between oxygen and HE2 atom of the CYS58, HIS57 (red colour) in the MMP12 and an oxygen and H atom of rutin.
Discussion
Inhibition of matrix metalloproteinase might be a valuable approach for the treatment of chronic diseases. The flavonoid and amino acid composite are thermodynamically stable. However, the binding affinity varied depends upon the hydrophilic and aromatic amino acid residues. The amino acid residues from elastase such as tryptophan (TRP), glutamine (GLU), lysine (LYS), Phenylalanine (PHE), glutamic acid (GLU), aspartic acid (ASP), arginine (ARG) and Histidine (HIS) are higher affinity to flavonoid catechin [18]. In the present study, rutin showed interaction with elastase by the involvement of alanine, cysteine and Histidine amino acid residues. It might be increase the rate of wound strength by increasing the elastin content.
Collagenase are zinc dependent endopeptidase which causes bronchitis inflammation in between the nose and lungs [19]. It is also a novel modulator of inflammation during sepsis. Recently, Saragusti et al., [20] reported the quercetin derivatives possess active interaction with the MMP-9 by their hydroxyl substitutions such as R3-OH, R4-OH or R5-OH of chromone. Inhibition of elastase might be helpful for embryonic development, sexual reproduction, and tissue remodelling. Interestingly, Almeida et al., [21] proved the wound healing potential of rutin-hydrogel in animal model. Gelatinase is a zinc dependent endopeptidase involved in extracellular matrix gelatin and type IV, type V, type VII, type IX and type X collagen degradation. The gelatinase combined with other matrix metalloproteinase and involved in ovulation, embryogenesis, membrane gland involution, bone development and wound healing [22]. The elevated expression of gelatinase A with gelatinase B enzyme leads to ovarian, retinoblastoma and hepatocellular carcinoma [23]. The increased expression leads to prognosis, glioma invasion and breakdown of basement membrane degradation, and tumour vasculature [24]. In the present findings, rutin exhibited highest binding affinity with MMP-2 which reflects the possible role in pathophysiology of malignant gliomas. A member of the metalloproteinase gene family is stromelysin, which plays critical role in connective tissues development. The elevated level of stromelysin in patient’s serum and synovial fluids causes bone joint diseases including rheumatoid arthritis, osteoarthritis and intracranial aneurysms cerebrovascular disorders [25]. Rutin with MMP8 showed ALA184, ALA182, HIS222, HIS218 and ALA184 hydrogen bond donor, which might be successfully involved in decreased the collagenase and increases the collagen content of extracellular matrix [25].
Conclusion
In conclusion, ruin showed binding affinity to four different classes of metalloproteinase. The domain structure of matrix metalloproteinase contain signal sequence, zinc-ligating thiol group and binding site, collagen binding fibronectin type II inserts and hinge region (Kundu and Patil, 2006). However, further study will be needed to find out the interaction of rutin with exact metallo protein domain regions. It would be greater possibilities to down regulate the action of metalloproteinase by the naturally occurring plant flavonoid like rutin.
Acknowledgements
The authors wish to thank the authorities of Annamalai University, Tamil Nadu, India for providing all support during the study period.