Research Article - (2018) Volume 4, Issue 2
Shihori Tanabe1*, Kazuhiko Aoyagi2, Hiroshi Yokozaki3 and Hiroki Sasaki2
1Division of Risk Assessment, Biological Safety Research Center, National Institute of Health Sciences, Kawasaki, Japan
2Department of Translational Oncology, FIOC, National Cancer Center Research Institute, Tokyo, Japan
3Department of Pathology, Kobe University of Graduate School of Medicine, Kobe, Japan
*Corresponding Author:
Shihori Tanabe, PhD
Division of Risk Assessment, Biological Safety Research Center, National Institute of Health Sciences
3-25-26, Tonomachi, Kawasaki-ku, Kawasaki, 210-9501, Japan.
Tel: +81-44-270-6686
Fax: +81-44-270-6703
E-mail: stanabe@nihs.go.jp
Received date: March 15, 2018; Accepted date: April 17, 2018; Published date: April 25, 2018
Citation: Tanabe S, Aoyagi K, Yokozaki H, Sasaki H (2018) Molecular Network Pathway of ERBB In Diffuse-Type Gastric Cancer: Mesenchymal Stem Cells and Epithelial-Mesenchymal Transition. J Clin Epigenet 4:13. doi: 10.21767/2472-1158.100098
Epithelial-mesenchymal transition (EMT) is related to malignancy and metastasis in cancer. The molecular networks including tyrosine kinases are altered in gastric cancer (GC). This study aims to reveal the role of ERBBs (erb-b2 receptor tyrosine kinases) in EMT, and generate the molecular network pathway of ERBBs in diffusetype GC and mesenchymal stem cells (MSCs). The expression of ERBB genes was analyzed in MSCs and diffuse-type GC which has mesenchymal characteristics compared to intestinal-type GC. The signaling and molecular network of ERBB was analyzed using several databases, including cBioPortal for Cancer Genomics, Kyoto Encyclopedia of Genes and Genomes (KEGG), BioGRID and VaProS. ERBB2 and ERBB3 gene expression were up-regulated in diffuse-type GC compared to MSCs. The ERBB3 molecular network includes epidermal growth factor receptor (EGFR), cadherin 1 (CDH1), catenin beta 1 (CTNNB1) and EPH receptor A5 (EPHA5). These results demonstrate the importance of the ERBB network in cancer signaling, and revealed a ERBB3 network pathway model in diffuse-type GC and MSCs, and EMT.
Keywords
Epithelial-mesenchymal transition; Gastric cancer; Mesenchymal stem cell; ERBB; Stem cell
Introduction
An abundance of molecular signaling pathways are linked between stem cells and cancer. As cellular phenotype changes, expression levels of specific genes in these signaling pathways are altered. We have previously investigated CTNNB1 networks in epithelial-mesenchymal transition (EMT), and revealed that the CTNNB1 network includes epidermal growth factor receptor (EGFR) and erb-b2 receptor tyrosine kinase (ERBB2) [1]. The ERBB family includes EGFR (ERBB1 or HER1), ERBB2 (HER2), ERBB3 (HER3) and ERBB4 (HER4) [2]. Previous studies have demonstrated that ERBB family members are important for cancer, and so in this article, we analyzed the ERBB network [3-5]. ERBB2 and ERBB3 were found to be overexpressed in an appendiceal adenocarcinoma in which p53 was inactivated and the Ras/RAF/MEK, Akt/mTOR, Wnt, and NOTCH pathways were activated [6]. Somatic mutations in ERBB3 were identified in a significant number of early-onset sporadic diffuse-type gastric cancer (GC) [7]. Mutations in extracellular domains of ERBB2 are suggested to be oncogenic in lung cancer [8].
Since previous reports have demonstrated that the ERBB members are involved in cancer and stem cell networks, it is important to elucidate the role of ERBBs in EMT and cancer resistance. To further reveal the mechanism of EMT in cancer and stem cells, we investigated the expression of ERBB genes in mesenchymal stem cells (MSCs) and diffuse-type GC, and generated ERBB network pathway model.
Materials and Methods
Gene expression analysis of MSCs and GC
Gene expression in MSCs (6 early- and 6 late- stage cultures, n=12) and diffuse-type GC (n=5) was analyzed with GeneChip® Human Genome U133 Plus 2.0 microarray (Affymetrix, Santa Clara, California, USA), as previously described [1,9,10]. Briefly, total purified cellular RNA was biotinylated and hybridized to the microarray. The signal intensity for each gene was analyzed and compared between MSCs and diffuse-type GC. The microarray data for MSCs and diffuse-type GC are available to the public in the NCBI Gene Expression Omnibus (GEO) database and are accessible via GEO Series accession number GSE7888 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7888) and GSE42252 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42252), respectively [1,9,10].
Cell culture
Human MSCs from bone marrow (Lonza, Walkersville, MD, USA) were cultured in MSC growth medium (MSCGM; Lonza #PT- 3001; MSC basal medium supplemented with mesenchymal cell growth supplement, L-glutamine and penicillin/streptomycin) at 37°C in a CO2 (5%) incubator. Cells were passaged according to the manufacturer’s protocol, however a slight modification was made to use trypsin-EDTA solution (Lonza #CC-3232). Lot numbers of the human MSC batches were as follows: #4F1127, #4F0312, #5F0138, #4F1560, #4F0591 and #4F0760. Informed consent was obtained for Poietics human MSC systems (Lonza) [10]. Passage numbers of MSC cultures were #4 and #22 for #4F1127, #4 and #28 for #4F0312, #4 and #24 for #5F0138, #5 and #28 for #4F1560, #4 and #28 for #4F0591, and #4 and #28 for #4F0760.
Diffuse-Type GC tissues
Diffuse-type GC tissues were originally provided by the National Cancer Center Hospital after obtaining written informed consent from each patient and approval from the National Cancer Center Institutional Review Board. All cancer specimens were reviewed and classified histopathologically according to the Japanese Classification of Gastric Cancer. Following surgical extraction, tissue specimens were immediately frozen with liquid nitrogen and stored at -80°C until microarray analysis [1,9]. Existing, publicly available data were analyzed in the article.
Analysis with Cbioportal for Cancer Genomics
cBioPortal for Cancer Genomics (https://www.cbioportal.org) [11,12] was used to analyze cancer genomics data related to ERBB3. The term “ERBB3” was searched in the cBioPortal for Cancer Genomics, and the cross-cancer alteration summary for ERBB3 was obtained. Studies of stomach adenocarcinoma (TCGA, Nature 2014) were further analyzed for pathway network analysis [13].
Gene Ontology Analysis of ERBB3
Gene Ontology of ERBB3 was analyzed using several databases, including the DAVID (https://david.ncifcrf.gov/home.jsp), EMBLEBI (https://www.ebi.ac.uk/QuickGO/), AmiGO 2 (https://amigo.geneontology.org/amigo/landing) and the Gene Ontology Consortium (https://geneontology.org/).
Pathway Network Analysis of ERBB3 and Related Genes
Pathway network analysis was performed using the databases VaProS (https://pford.info/vapros/), KEGG (https://www.genome.jp/kegg/), cBioPortal for Cancer Genomics (https://www.cbioportal.org), and Cytoscape (https://www.cytoscape.org/). Molecular interactions were analyzed using the BioGRID (https://www.thebiogrid.org) database [14]. Molecule localization was analyzed using The Human Protein Atlas (https://www.proteinatlas.org/) [15] and UniProt (https://www.uniprot.org/) [16].
Statistical Analysis
The gene expression data was shown in signal intensity or ratio to the average of MSC expression. The ratio to the mean of signal intensity in MSC samples normalized with GAPDH signal intensity was calculated and averaged. The data were shown as the mean ± SE. Student’s t-test was performed in Microsoft Excel (Microsoft, Redmond, WA, USA). p<0.001 (n=12 in MSCs, n=5 in diffuse-type GC) was considered as statistically significant
Results
Gene expression of EGFR, ERBB2 and ERBB3 in MSCs and diffuse-type GC
Expression levels of the EGFR, ERBB2 and ERBB3 genes were analyzed in diffuse-type GC and MSCs (Figure 1A). ERBB2 gene expression was up-regulated approximately 3-fold in diffusetype GC compared to MSCs (Figure 1B). ERBB3 gene expression was up-regulated by approximately 150-fold in diffuse-type GC compared to MSCs (Figure 1C). Signal Intensity of the ERBB3 gene expression in each sample is shown in Figure 1D. ERBB4 signal intensity was low.
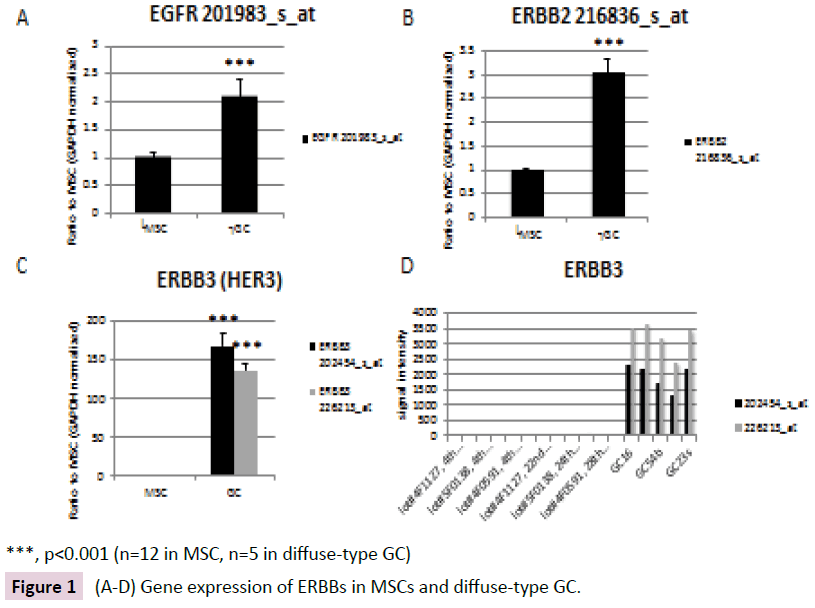
Figure 1: (A-D) Gene expression of ERBBs in MSCs and diffuse-type GC.
ERBB3 Interaction Analysis
ERBB3 interactions were analyzed using BioGRID database (Figure 2). ERBB3 interactors included growth factor receptor bound protein 2 (GRB2), phosphoinositide-3-kinase regulatory subunit 2 (PIK3R2), C-terminal Src kinase (CSK), SHC adaptor protein 1 (SHC1) and epidermal growth factor (EGF) [2,17].
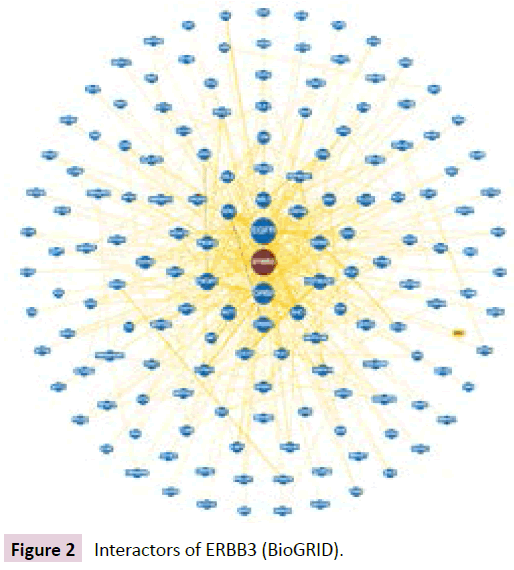
Figure 2: Interactors of ERBB3 (BioGRID).
Network Pathway Analysis of ERBB3
cBioPortal for Cancer Genomics network pathway analysis revealed the ERBB3-associated network and ERBB3 cross-cancer alterations (Figure 3A and B). The ERBB3 molecular network was analyzed in stomach adenocarcinoma (TCGA, Nature 2014, tumor samples with sequencing and CNA data 287 samples / 1 gene) [13]. The network contained 51 nodes and included the 1 query gene ERBB3 as well as the 50 most frequently altered neighboring genes (out of a total of 420) in cBioPortal (Table 1). The network shown in Figure 4 includes the 1 originally queried core gene and the 50 neighboring genes. Analysis of molecular interactions in VaProS demonstrated that ERBB3 interacts with neuregulin 1 (NRG1) and SH2B adaptor protein 3 (SH2B3).
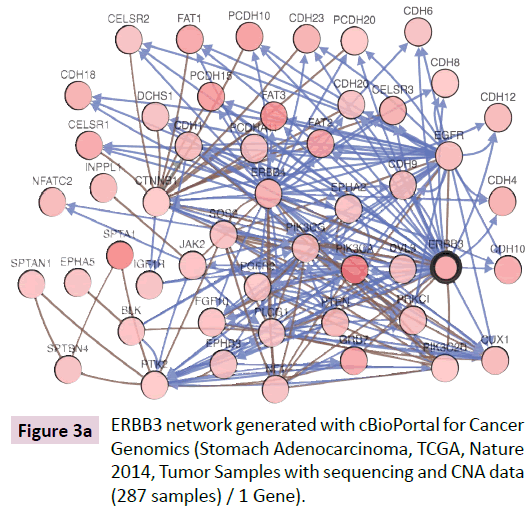
Figure 3a: ERBB3 network generated with cBioPortal for Cancer Genomics (Stomach Adenocarcinoma, TCGA, Nature 2014, Tumor Samples with sequencing and CNA data (287 samples) / 1 Gene).
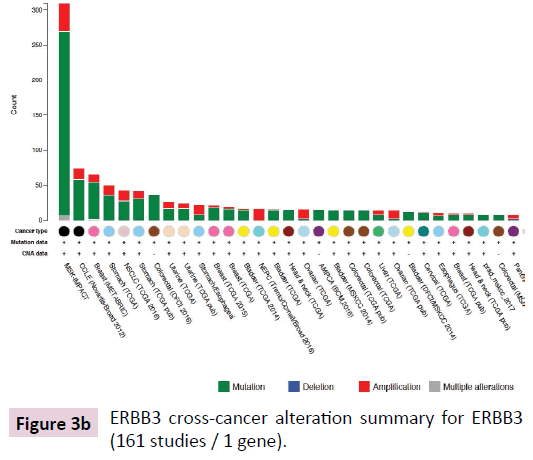
Figure 3b: ERBB3 cross-cancer alteration summary for ERBB3 (161 studies / 1 gene).
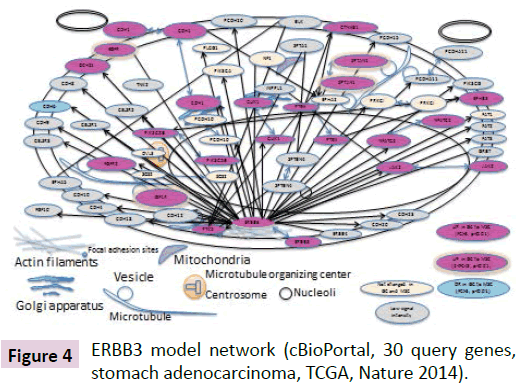
Figure 4: ERBB3 model network (cBioPortal, 30 query genes, stomach adenocarcinoma, TCGA, Nature 2014).
| Gene Symbol | Gene Title | Entrez Gene | Probe Set ID |
|---|---|---|---|
| BLK | BLK proto-oncogene, Src family tyrosine kinase | 640 | 206255_at |
| CDH1 | cadherin 1 | 999 | 201131_s_at |
| CDH10 | cadherin 10 | 1008 | 220115_s_at |
| CDH12 | cadherin 12 | 1010 | 207149_at |
| CDH18 | cadherin 18 | 1016 | 206280_at |
| CDH20 | cadherin 20 | 28316 | 210913_at |
| CDH23 | cadherin related 23 | 100653137 / 64072 | 224527_at |
| CDH4 | cadherin 4 | 1002 | 239485_at |
| CDH6 | cadherin 6 | 1004 | 205532_s_at |
| CDH8 | cadherin 8 | 1006 | 217574_at |
| CDH9 | cadherin 9 | 1007 | 207729_at |
| CELSR1 | cadherin EGF LAG seven-pass G-type receptor 1 | 9620 | 41660_at |
| CELSR2 | cadherin EGF LAG seven-pass G-type receptor 2 | 1952 | 36499_at |
| CELSR3 | cadherin EGF LAG seven-pass G-type receptor 3 | 1951 | 40020_at |
| CTNNB1 | catenin beta 1 | 1499 | 223679_at |
| CUX1 | cut like homeobox 1 | 1523 | 225935_at |
| DCHS1 | dachsous cadherin-related 1 | 8642 | 222101_s_at |
| DVL3 | dishevelled segment polarity protein 3 | 1857 | 201908_at |
| EGFR | epidermal growth factor receptor | 1956 | 201983_s_at |
| EPHA2 | EPH receptor A2 | 1969 | 203499_at |
| EPHA5 | EPH receptor A5 | 2044 | 237939_at |
| EPHB3 | EPH receptor B3 | 2049 | 204600_at |
| ERBB3 | erb-b2 receptor tyrosine kinase 3 | 2065 | 202454_s_at |
| ERBB4 | erb-b2 receptor tyrosine kinase 4 | 2066 | 214053_at |
| FAT1 | FAT atypical cadherin 1 | 2195 | 201579_at |
| FAT2 | FAT atypical cadherin 2 | 2196 | 208153_s_at |
| FAT3 | FAT atypical cadherin 3 | 120114 | 236029_at |
| FGF10 | fibroblast growth factor 10 | 2255 | 231762_at |
| FGFR2 | fibroblast growth factor receptor 2 | 2263 | 208228_s_at |
| GRB7 | growth factor receptor bound protein 7 | 2886 | 210761_s_at |
| IGF1R | insulin like growth factor 1 receptor | 3480 | 225330_at |
| INPPL1 | inositol polyphosphate phosphatase like 1 | 3636 | 201598_s_at |
| JAK2 | Janus kinase 2 | 3717 | 205841_at |
| NF1 | neurofibromin 1 | 4763 | 212676_at |
| NFATC2 | nuclear factor of activated T-cells 2 | 4773 | 228442_at |
| PCDH10 | protocadherin 10 | 57575 | 228635_at |
| PCDH15 | protocadherin related 15 | 65217 | 1553344_at |
| PCDH20 | protocadherin 20 | 64881 | 232054_at |
| PCDHA11 | protocadherin alpha 11 | 56134 / 56135 / 56136 / 56137 / 56138 / 56139 / 56140 / 56141 / 56142 / 56143 / 56144 / 56145 / 56146 / 56147 / 9752 | 210674_s_at |
| PIK3C2B | phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta | 5287 | 204484_at |
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha | 5290 | 204369_at |
| PIK3CG | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma | 5294 | 206369_s_at |
| PLCG1 | phospholipase C gamma 1 | 5335 | 202789_at |
| PRKCI | protein kinase C iota | 5584 | 209678_s_at |
| PTEN | phosphatase and tensin homolog | 5728 | 233254_x_at |
| PTK2 | protein tyrosine kinase 2 | 5747 | 1559529_at |
| SOS2 | SOS Ras/Rho guanine nucleotide exchange factor 2 | 6655 | 212870_at |
| SPTA1 | spectrin alpha, erythrocytic 1 | 6708 | 206937_at |
| SPTAN1 | spectrin alpha, non-erythrocytic 1 | 6709 | 208611_s_at |
| SPTBN4 | spectrin beta, non-erythrocytic 4 | 57731 | 224551_s_at |
| TNK2 | tyrosine kinase non receptor 2 | 10188 | 203839_s_at |
Table 1: Molecules in ERBB3 network.
Model Network of ERBB3
The model ERBB3 interaction network is shown in Figure 4. Nodes are colored based on gene expression alteration in diffuse-type GC compared to MSCs. Pink nodes indicate up-regulated (UR) genes and blue nodes indicate down-regulated (DR) genes with FC>3, p value <0.01. UR genes with 2
Primary localization information was based on the Cell Atlas in the Human Protein Atlas where available. Secondary localization information was based on subcellular location information from the UniProt database. Gene ontology (GO) of molecules in the ERBB3 model network was related to Wnt receptor signaling pathway, cell adhesion, cell proliferation, cell differentiation and apoptosis.
KEGG pathway and Gene Ontology of ERBB3
According to KEGG pathway analysis, ERBB3 is involved with EGFR tyrosine kinase inhibitor resistance, the ErbB signaling pathway, the calcium signaling pathway, endocytosis, proteoglycans in cancer, and microRNAs in cancer. The GO of ERBB3 analyzed in AmiGO 2 includes the ERBB3 signaling pathway, the ERBB4-ERBB3 complex, the ERBB2-ERBB3 signaling pathway, and regulation of the ERBB3 signaling pathway (https://amigo.geneontology.org/amigo/search/ontology?q=ERBB3). GO biological process of ERBB3 included transmembrane receptor protein tyrosine kinase signaling pathway and regulation of cell proliferation.
DAVID analysis of ERBB3 network molecules
Molecules of ERBB3 network were analyzed in Functional Annotation Tool of DAVID Bioinformatics Resources 6.8, NIAID/ NIH (https://david.ncifcrf.gov/home.jsp). The Gene Ontology and Online Mendelian Inheritance in Man (OMIM) disease for the molecules of ERBB3 network were analyzed. Molecules in ERBB3 network are involved in cell adhesion or mitogen-activated protein kinase (MAPK) cascade.
Enrichment Analysis of ERBB3
The ERBB3 enrichment analysis using cBioPortal for Cancer Genomics revealed that the mRNA expression levels of DLG associated protein 3 (DLGAP3) and RNA polymerase II subunit J2 (POLR2J2) were altered in ERBB3-altered cases and ERBB3- unaltered cases in stomach adenocarcinoma (TCGA, Nature 2014) [13].
Discussion
In this study, we investigated the ERBB signaling role in EMT and cancer. Gene expression of ERRB3, as well as ERBB2, was upregulated in diffuse-type GC compared to MSCs. ERBB3 network pathway model generated in this study included the EMT-related molecules such as CTNNB1 and CDH1. ERBB3 plays an important role in the PI3K/AKT pathway and is thus a target for the treatment of solid tumors [18]. Cell survival can be inhibited by a mouse monoclonal antibody against ERBB3 [18]. Expression of ERBB3 is correlated with intestinal stem cell markers such as LGR5, EPHB2, CD44 in colorectal cancer, although ERBB3 expression is observed in the differentiated cell population at the top of the crypts [19]. Furthermore, ERBB3 is targeted by miR- 205-5p, a tumor suppressor in breast cancer [20]. Knockdown of ERBB3 suppressed migration and invasion of bladder cancer cells [21]. miR-148a-3p targets both ERBB3 and AKT2 and may repress ERBB3- and AKT-mediated proliferation and EMT [21]. ERBB3 is characterized as a marker for circulating tumor cells exhibiting EMT phenotype and therapeutic resistance [22]. The activation of EGF/EGFR-ErbB2 and NRG1/ErbB2-ErbB3 autocrine signaling loops drives triple negative breast cancer exhibiting the chemoresistance [23]. It seems that ErbB3 receptor is important for perineural migration to the bone marrow [24]. The previous immunohistopathology investigation showed that the ERBB2 and ERBB3 proteins were overexpressed in GC [25]. The previous finding indicating the correlation with ERBB3 overexpression and overall survival in GC is consistent with the present discussion in which ERBB3 up-regulation may be related to malignancy or the EMT network in GC [25]. A report suggests that ERBB2 and ERBB3 are down-regulated by DNA-damaging agent such as cisplatin via ERK and p38 signaling pathway [26]. It is also demonstrated that ERBB3 is involved in resistance to EGFR or ERBB2 targeted therapy for cancer [27]. Furthermore, the study of nanoparticles targeting ERBB2 and microRNA21 suggests the importance of ERBB signaling in cancer [28]. These insights indicate the significance of regulation of ERBB2 and ERBB3 signaling in cancer.
The down-regulation of ERBB3 following EMT leads to a reduction in serum-independent proliferation and an enhancement of PI3K-dependent proliferation [29]. Molecules related to intercellular adhesion and metastatic processes (such as PTEN) and endocytosis (such as ERBB3) are involved in liver metastasis [30]. In ERBB2-positive cancers, expression of EGFR, ERBB3 and CDKN1B are demonstrated to predict and distinguish the PI3K and MAPK signaling axes [31]. The ERBBs are inhibited by N-myc downstream regulated gene 1 (NDRG1) [32]. Since ERBBs are involved in oncogenic pathways, such as the PI3K, Akt and STAT3 pathways, further investigation of targeted molecules in the ERBB network is needed. Considering that the cancer cells migrating and metastasizing exhibit EMT-like phenotype, the investigation of ERBB3 signaling in bone-marrow derived MSCs showing mesenchymal phenotype and diffuse-type GC that exhibits more EMT-like phenotype compared to intestinal-type GC may lead to the novel discoveries for the overcome of the therapeutic resistance. Although the current finding showing ERBB3 gene up-regulation in diffuse-type GC compared to MSCs needs to be further examined in terms of the discussion whether the ERBB3 plays crucial role in EMT or not, it is suggested that the ERBB3 signaling is activated in gastric cancer. To further investigate the role of ERBB3 signaling in EMT and CSCs, the analysis in diffusetype GC and intestinal-type GC would be needed in future.
In conclusion, ERBB3 gene expression was up-regulated in diffuse-type GC compared to MSCs, and ERBB3 network pathway model in EMT was generated. The model of the ERBB3-related molecular network included the EMT-associated molecules such as CTNNB1 and CDH1.
Acknowledgement
We gratefully acknowledge Dr Ryoji Kushima for the pathological and clinical evaluations. We would also like to thank Ms. Rie Komatsuzaki and Ms. Fumiko Chiwaki for their technical assistance. This study was supported in part by the National Institute of Biomedical Innovation, the Ministry of Health, Labour and Welfare of Japan, National Cancer Center Research and Development Fund, the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Princess Takamatsu Cancer Research Fund. The authors would like to thank Dr. Akihiko Hirose and Dr. Takashi Yamada for supporting the preparation of the manuscript. The authors would like to thank Prof. Kei Yura for introducing us to the VaProS database, which was used in part in this research. VaProS is a data-cloud developed by the Information Core of the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and Development (AMED).