Research Article - (2022) Volume 12, Issue 11
Received: 17-May-2022, Manuscript No. EJEBAU-22-13482; Editor assigned: 19-May-2022, Pre QC No. EJEBAU-22-13482(PQ); Reviewed: 02-Jun-2022, QC No. EJEBAU-22-13482; Revised: 10-Oct-2022, Manuscript No. EJEBAU-22-13482(R); Published: 17-Oct-2022, DOI: 10.36648/2248-9215.12.82
Gnetum africanum is a twisted woody climbing wild plant liana found growing naturally in dense equatorial rainforest but currently domesticated in tropical areas of the world. There has been taxonomic confusion in classifying this liana due to its observable diverse morphological characters. Indeed, morphological characterization alone has not offer the needed solution. However, there is dearth of information on the molecular characterization of Gnetum africanum. Morphological and molecular descriptors, could help offers the needed phenotypic and genotypic characterization of this plant much better. This study is aimed at characterizing Gnetum africanum land races to identify species that could be used for the development of high yielding hybrids. Three different land races or accessions of Gnetum africanum was collected from different locations in Nigeria. High quality genomic DNA was isolated from the different samples; it was checked for purity and quantified as appropriate. The DNA samples were subjected to PCR amplification and DNA barcoding studies (chloroplast large subunit of ribose 1,5-bisphophate carboxylase rbcL gene). The amplicons were gel eluted, sequenced and checked for homology by using Basic local alignment search tool BLAST. Identification was obtained from the top similar outcomes of the samples. For Ekim (1b) sample, the forward blast showed 95% similarity with Basella alba voucher ID: NC041293.1 and the reverse had 88% similarity with Talinum fruticosum cultivar ID: MK598685.1. For etinan (1a) sample, the forward blast had 86% similarity with rhabdo thamnus solandri voucher ID: JQ933464.1 and for reverse, there was no similarity founded.
Gnetum africanum; Germplasm; DNA barcoding; BLAST; Molecular characterization
Gnetum africanum welw is a shade loving edible climbing plant widely used as leafy vegetable and grows naturally in dense equatorial rainforest of Africa. It comprises of approximately 50 species of climbing plants Liana distributed in the tropical areas of the world. Gnetum africanum are dioeciously vines occurring only in the African continent. These species are one of the most popular green leafy vegetables in Nigeria, and is equally gaining popularity as a delicious vegetable across other African countries as Angola, Cameroon, Central African Republic, Congo, Equatorial Guinea, Gabon and democratic republic of Congo for instance, in Nigeria, it is called afang (Efik/Ibibio), ukazi (Igbo), yala (Ogoja), ajaabaje, ajakotale (Yoruba). In Cameroon, it is called eru, okok, mfumbua or fumbua and in Angola, Gabon, Central Africa Republic, it is called koko. The rich diversity of the indigenous leafy vegetables of Nigeria has been documented by several researchers including, but there has been little or no work done on Gnetum africanum such as in depth genetically and morphological characterization or controlled hybridization of the said plant species. It’s obvious that taxonomic confusion, coupled with the limited knowledge regarding genetic and geographical differentiation of this plant species have limited domestication, conservation and utilization of Gnetum africanum. However, knowledge of the morphological or genetic diversity existing among the available Gnetum africanum land races are important prerequisite for preliminary characterization of this leafy vegetable. In this era of biotechnology, genetic diversity studies involving characterization of Germplasm features the use of molecular markers such as Polymerase chain reaction PCR, Amplified Fragment Length Polymorphisms (AFLP), Random Amplified Polymorphic DNA (RAPD), Inter Sequence Simple Repeats (ISSR) and DNA based marker system. The concept of morphological characterization is very important in plant improvement, especially when it is desired to study and compare the performance of each specie and in hybrids combination. The study was aimed at characterizing Gnetum africanum land races to identify desirable characters in the accession that could be used for the development of high yielding hybrids for domestication [1-4].
Samples Collection
Three different Samples of Gnetum africanum were collected in February 2021, across different locations in Nigeria. The fresh leaves of the different samples were used for DNA extraction and subsequent characterization purpose. They were preserved in silica gel which was safely transported to the research laboratory/center (International Institute of Tropical Agriculture (IITA)), Oyo State, Nigeria for further study [5].
Morphological Description of Gnetum africanum
To analyze the morphological variation among the three accessions of the collected Gnetum africanum plant, a twig of young leaves were sampled from each accession and crossed examined according to the method. The visual examination was performed to detect any morphological differences among the samples, with respect to the leaf size, shape and colour. Differences in the floral structure, inflorescence types, peduncle, growth habit and style were also factored in to serve similar purpose in the morphological characterization according to Nya and Eka [6].
Statistical Analysis
The study were performed with a minimum of four replications per treatment and the data were analyzed statistically using SPSS statistics 19. The mean values were expressed as mean ± SE and the significant differences among means was carried out at 5% probability level using Duncan's Multiple Range Test (DMRT) [7].
Molecular Characterization of Gnetum Africanum
Preparation of 50 ml N-2-Hydroxyethylpiperazine-N-2-Ethane sulfonic acid (HEPA) buffer solution for DNA isolation. To prepare an HEPES buffer solution, analytical balance was used to weigh 1.94 g of L-ascorbic acid, 2 g of Poly Vinyl Pyrrolidone (PVP) into a conical flask and 180 ml of distilled water was added. 20 ml of HEPES buffer and 1 ml of β-mercaptoethanol was added and stirred to dissolve using magnetic stirrer [8].
Preparation of Chloroform Isoamyl Alcohol (CIA) solution
24 ml of chloroform was measured using a measuring cylinder and poured into a beaker and 1 ml of isoamyl alcohol was added and mixed together in the beaker to obtain the CIA solution [9].
Preparation of 70% Ethanol for Extraction Purification
70% ethanol was prepared according to Nya [10].
Preparation of Cetyl Trimethyl Ammonium Bromide (CTAB) extraction Buffer
1 g of Poly Vinyl Pyrolidone (PVP) was weighed into a conical flask containing 2 g of Cetyl Trimethyl Ammonium Bromide (CTAB), 10 ml of 1 M tris hydrogen chloride and 5 ml of 0.5 M Ethylene Diamine Tetracetic Acid (EDTA) was also added into the flask before 82 ml of distilled water was added and the solution was stirred till a foamy solution was formed [11].
DNA extraction from Gnetum africanum (afang)
Total genomic DNA of these accessions was extracted from leaf samples following a modification of the Cetyl Trimethyl Ammonium Bromide CTAB extraction protocol with RNAse A treatment. The collected Gnetum africanum samples were stored in silica gel and safely transported to the research laboratory of (International Institute of Tropical Agriculture (IITA)) where it was kept at -86°C upon arrival [12].
Lysis of Sample’s Cell Wall
Eppendorf tubes were labeled in 4 replicates and left on ice for total genomic DNA isolation. To lyse the samples, liquid nitrogen was used to freeze dry the samples for grinding. The grinded samples were collected into another Eppendorf tubes and place on ice [13].
Separation Process
1 ml of HEPES (4-(2-hydroxyethyl)-1-piperazin ethane sulfonic acid) buffer was added to the grinded samples in eppendorf tubes and tap for few min to mixed. Then it was centrifuged at 1000 rpm for 10 min at the temperature of 11°C. After centrifugation, the supernatant was decanted and discarded while the pellet was left. The process was repeated twice [14].
DNA Purification
For this, 70 μl of Cetyl Trimethyl Ammonium Bromide CTAB was added to the DNA pellet and tapped to mix and then incubated in water bath for 30 min while inverting it at 10 min intervals till the 30 min was completed. The samples were brought out and left on the fume hood to cool. 600 μl of Chloroform Isoamyl Alcohol (CIA) was added to the samples and shaken for 10 min to mix and was centrifuged at 1000 rpm for 10 min at 4°C temperature. The supernatant was collected into clean new tubes while the other part (pellet) was discarded; the CIA wash process was repeated once again [15].
DNA Precipitation
The supernatant was transferred into fresh labeled tubes, 800 μl of ice cold isopropanol was added and inverted gently for 2 min and stored in -80°C for 30 min. Thereafter, it was brought out and spin at 35000 rpm for 30 min, pellet was formed and the supernatant was discarded. 400 μl of 70% ethanol was added to the pellet and spin again at 10000 rpm for 10 mins, the process was repeated for one other time and the supernatant was discarded and the extracted DNA was left on bench to air dry for 30 min. 50 μl of RNAse free water (elution buffer) was added to dissolve the precipitated DNA and remove any trace of RNA remains [16].
DNA Quality Analysis on Gel Electrophoresis
Agarose gel preparation: The quality of the DNA samples was assessed by agarose gel electrophoresis. Briefly, 0.8 g of agarose was weighed into 100 ml of 1 x TBE in a conical flask.
The solution was allowed to dissolve completely in the microwave for 3 min. It was later placed in a water bath to cool at 60°C for a min. 5 μl of ethidium bromide was added and gently swung to mix. Then, it was poured into the gel tray/caster which served as a mold and was allowed to solidify in 20 min. The solidified gel was submerged with a TBE buffer filled in electrophoresis chamber with its comb inserted, the negative and positive poles connected to the power source.
The comb was removed gently and the samples were loaded into the wells [17].
Samples preparation for loading: Samples were prepared for electrophoresis by mixing it with loading dye at same density.
This made the samples to sink through the buffer and remains in the wells and the samples were delivered into the wells with the use of micropipette. 3 μl of DNA and 3 μl of loading dye was pipette into a reaction plate to mix. 6 μl of the mixture was loaded into the wells of the prepared agarose gel in the electrophoresis chamber. The gel was left to run at 100 volts for about 45 min. Thereafter, the sample fragments were visualized via an FLA-5100 imaging system (Aplegen- Eduro GDS gel documentation Beckman system) with a resolution of 50 mm.
DNA Quanti ication Analysis on Nano Dr op Spectrophotometer
The DNA concentration was determined using nano drop ND-1000 spectrophotometer (nano drop technologies, Wilmington, USA) program. The nano drop program was opened and clicked on the blank. The blank was loaded with elution buffer that was used to suspend the DNA. The upper and lower pedestal was wiped using Kim, then 2 μl of the DNA samples were loaded on the lower measurement pedestal and was closed with upper pedestal carefully to avoid bubble.
Thereafter, the measurement bottom was click on and the reading was taken, it was repeated till other samples reading were completely taken.
DNA barcoding study of the different land races of Gnetum africanum
To investigate the genetic variability among the three different accessions of Gnetum africanum samples, DNA barcoding studies was conducted using ribulose-1,5- bisphosphate carboxylase (rbcL) chloroplast barcode. PCR amplification of the most frequently used barcodes two coding regions (rbcL and matK) of the chloroplast and total genomes was performed. Primers from rbcL gene sequences used for PCR amplification is shown on the Table 1 below.
| Name | Primer code and length | Sequence(5’-3’) |
|---|---|---|
| rbcL primers | ||
| Forward | RbcL F535 | CTTTCCAAGGCCCGCCTCA |
| Reverse | RbcL R705 | CATCATCTTTGGTAAAATCAAGTCCA |
Table 1: Sequence information of the forward and reverse primers of barcode used for the rbcL DNA barcoding studies.
The PCR reaction was carried out in a volume of 25 μl reaction mixture consisting of the following components: genomic DNA (100 ng/μl), taq DNA polymerase (10 x), rbcL primer (5p Mol), dNTPs (2.5 mM), Mgcl2 (50 mM), taq buffer (5 μ/μl), DMSO and a negative control containing nuclease free Milli-Q water was included to ensure there were no false positive cross or contamination, and a PCR reactions mixture was prepared as detailed on Tables 2 and 3.
| S/N | Component | Working concentration | Volume required per 25 µl reaction | Volume of cocktail x 4 |
|---|---|---|---|---|
| 1 | Genomic DNA | 3 µl | 3.0 µl | 3 µl each to make up |
| 2 | Reaction buffer | 5 µ/µl | 2.5 µl | 10.0 µL |
| 3 | DNTPs | 2.5 µl | 2.0 µl | 8.0 µL |
| 4 | Forward primer | 5p Mol | 1.0 µl | 4.0 µL |
| 5 | Reverse primer | 5p Mol | 1.0 µl | 4.0 µL |
| 6 | Taq DNA polymerase | 10 x | 0.1 µl | 0.4 µL |
| 7 | MgCl2 | 50 mM | 1.0 µl | 4.0 µL |
| 8 | DMOS | 1.0 µl | 4.0 µL | |
| 9 | Nucleases water | 13.4 µl | 53.6 µL | |
| Total | 25 µl | 88 µL |
Table 2: PCR reaction mixture prepared for the four different barcoding loci of Gnetum africanum.
| Initial denaturation cycling reaction | 94°C for 5 mins | |
|---|---|---|
| Denaturation | 94°C for 15 mins | 35 cycles |
| Annealing | 65°° for 20s | |
| Extension | 72°C for 30s | |
| Final Extension | 72°C for 7mins |
Table 3: PCR amplification conditions of the barcode of Gnetum africanum as carried out in a DNA thermal cycler (Gene Amp PCR system 9700).
After initial denaturation stage at 94°C for 5 min, thermo cycling was performed at 94°C for 15 s, 65°C for 20 s and 72°C for 30 s for 35 cycles with a final temperature at 72°c for 7 min then hold temperature 10°C for infinity. The primer was tested using genomic DNA from all the accessions. The PCR products were separated by electrophoresis at 90V for 1 hr 30 min in a 1.5% agarose gel (resolute wide range, BIOzym) with 1 x TBE electrophoresis buffer. DNA ladder (50 bp-1000 bp) was used as DNA marker. After a successful PCR was carried out, agarose gel electrophoresis analysis was also carried out.
Gels were stained with ethidium bromide and scanned using an FLA-5100 imaging system (Aplegen-Eduro GDS gel documentation Beckman system) with a resolution of 50 mm.
DNA Sequencing of the Amplified Product and Bioinformatics
The double stranded PCR products were purified using a PCR purification kit and directly sequenced from both the ends using dye terminator technique. Forward and reverse cycle sequencing based on the sanger’s sequencing method was performed using the big dye terminator v3.1 cycle sequencing kit (perkin-elmer, applied bio systems) on AB 3500 genetic analyzer (perkin-elmer, applied bio systems USA). The sequencing reaction mixture (20 μl) contained 2.5 x of ready reaction premix, 5 x of big dye sequencing buffer, 50 mg template (PCR product), 10 μM primer, deionized water (Table 4).
| Reagent | Volume |
|---|---|
| Ready reaction premix | 4.0 µL |
| Template DNA | 3 µL |
| Primer | 3.2 pmol |
| Deionized water | q.s (add up to 20 µL) |
| Big dye sequencing buffer | 2 µL |
| Final volume | 20 µL |
Table 4: Sequence reaction mixtures.
For each sample reactions, the above reagent was added to a separate tube mixed well and spinned briefly. It was used on a gene amp PCR system 9700 dual 384 well sample block module. The tubes were placed in a thermal cycler and set to the correct volume. An initial denaturation was performed, rapid thermal ramp 96°C for 1 min. The procedure was repeated for 25 cycles: rapid thermal ramp* to 96°C for 10 sec, rapid thermal ramp to 50°C for 5 sec, rapid thermal ramp to 60°C for 4 min, rapid thermal ramp to 4°C and hold until it was ready for purification, the 96 well reaction plate was removed from the thermal cycler then contents of the tubes were spinned down briefly using a micro centrifuge. 5 μL of 125 mM EDTA was added to each well. 60 μL of 100% ethanol was added to each well. The plate was sealed with aluminum tape and mix by inverting 4 times. It was incubated at room temperature for 15 min. Using a Coulter-AllegraTM 25R centrifuge, a plate adapter was used and spinned at a maximum speed of 1400-2000 mg for 45 min. The plate was inverted and spinned up at 185 mg, and then removed from the centrifuge. 60 μL of 70% ethanol was added to each well.
With the centrifuge set to 4°C, it was spinned at 1650 x g for 15 min. The plate was inverted and spinned up at 185 x g for 1 min, and then removed from the centrifuge. To continue, the samples were suspended in injection buffer. To store, it was covered with aluminum foil and stored at 4°C. Each of the samples was sequenced in the sense and antisense direction and analyzed with the AB sequence navigator software (perkin-elmer, Applied Bio systems). The nucleotide sequences of both DNA strands were obtained and compared to ensure accuracy. The sequence comparison to set up the level of identification (species, genus, or family) through Blastn algorithms. The nucleotide sequences obtained were used as queries in the Blastn search for molecular based species identification matches with the available GenBank sequences.
Morphological Description of Gnetum Africanum
Gnetum africanum plants that were collected from different locations within Akwaibom state were examined for morphological variation. A twig of young leaves collected from the parent plant at each locality was visually examined, photographed and documented. Morphologically, most of the plants showed distinct variation with respect to the leaf size, shape, color etc. The plant obtained from etinan appeared healthy with big round vined leaves which were succulent in nature. Ekim had smaller long leaves which were dark green in color. While a light green with much smaller round leaves was observed in the samples gotten from ikotakpaden making the sample look much tender in nature (Figure 1).
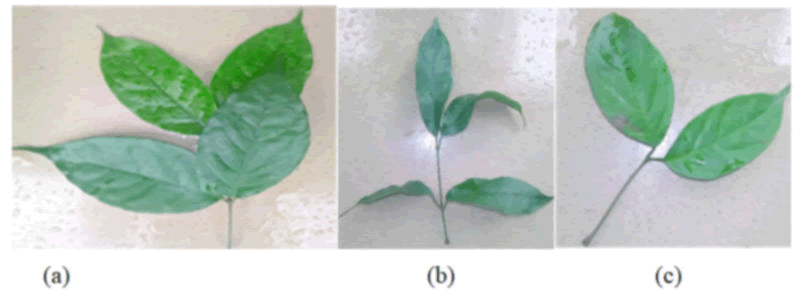
Figure 1: Morphological variation among the 3 different samples of Gnetum africanum as observed with the naked eye. The plant samples were collected from the following locations within Akwaibom state: (a) Etinan; (b) Ekim; (c) Ikotakpaden.
Molecular Characterization
DNA quality analysis on gel electrophoresis: The DNA extracts from the samples were analyzed in a 0.8% agarose gel with 1 x TBE buffer, for bands purity estimation under UVlight and documented using the gel documentation equipment (Aplegen-Eduro GDS gel documentation Beckman system). 50 bp DNA ladder (New England Bio labs Inc., UK) (50-1000 bp) was used as the DNA marker, the DNA bands were marked at 390 bp of the DNA ladder.
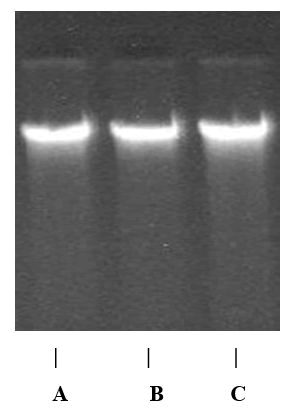
Figure 2: DNA gel image of the 3 different samples of Gnetumafricanum as observed with the gel documentation equipment (Aplegen-Eduro GDS Gel documentation Beckman system). From the left, the first gel image is sample collected from (A) Etinan, followed by the ones collected at; (B) Ekim and then the ones collected at; (C) Ikotakpaden with the DNA bands marked at 390bp of the DNA ladder.
DNA quantity analysis on nano drop spectrophotometer: The size of the DNA extract from each samples were checked using nano drop spectrophotometer with elution buffer loaded as blank for DNA quantity analysis. The result turns out great, in the Table 5 below.
| S/N | Sample name | Sample record | DNA conc. with unit | 260/280 |
|---|---|---|---|---|
| 1 | Etinan | A | 1399.6 mg/µl | 1.84 |
| 2 | Ekim | B | 513.8 mg/µl | 1.97 |
| 3 | Ikotakpaden | C | 1127.8 mg/µl | 1.88 |
Table 5: DNA quantity analysis.
PCR Amplification Analysis of Ribulose I, 5- Bisphosphate Carboxylase (Rbcl) Gene
With a 25 μl reaction mixture, the DNA barcode program was carried out as described and the PCR amplification was carried out in a DNA thermal cycler as per the detailed conditions. The chloroplast barcode was used for the DNA bar coding studies among 3 Gnetum africanum accessions.
Results of the test within the three samples with the rbcL barcode showed prominent PCR ampli ication with 100% success rate. The RBCL gene which codes for the large subunit of ribulose I, 5-bisphosphate carboxylase oxygenate (RuBisCo) enzyme is commonly used for molecular discrimination of plant species. This coding region was ampli ied for all the samples and amplicons of size 330bp was obtained in all the bands (Figure 3).
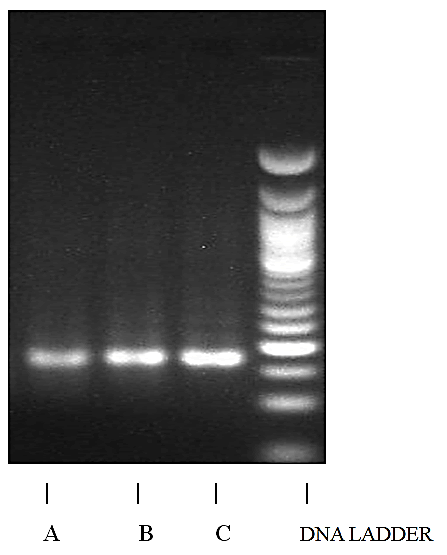
Figure 3: rbcL DNA barcoding profile for the three accessions of Gnetum africanum. From the left, the first gel image is a sample collected from (a) Etinan, followed by the ones collected at; (b)Ekim and then the ones collected at; (c)Ikotakpaden with the amplicons sizes marked at 330bp band of the ladder.
DNA Sequence of the Amplified Product and Bioinformatics
The amplicons obtained from the barcode was further sequenced with both forward and reverse primer (rbcL gene) and the nucleotide sequence obtained were not aligned at both ends. DNA sequence length of 535 bp for the rbcL forward sequence and 705 bp for reverse sequence length respectively. The sequence information was uploaded into the GenBank database and this ID’s was obtained from the top similar outcomes. For Ekim sample, Basella alba voucher with for the forward blast with 95% similarity and Talinum fruticosum cultivar with ID: MK598685.1, for the reverse with 88% similarity; For Etinan sample, rhabdo thamnussolandri voucher with ID: JQ933464.1 for forward with 86% similarity and for reverse, there was no similarities founded; For IkotAkpaden sample, there was no similarities founded for the forward sequence blast and alternanthraficoidea voucher with ID: MK757193.1 sequence ID for reverse with 95% sequence similarity respectively. The forward and reverse sequence of Gnetum africanum samples (Figures 4-9).
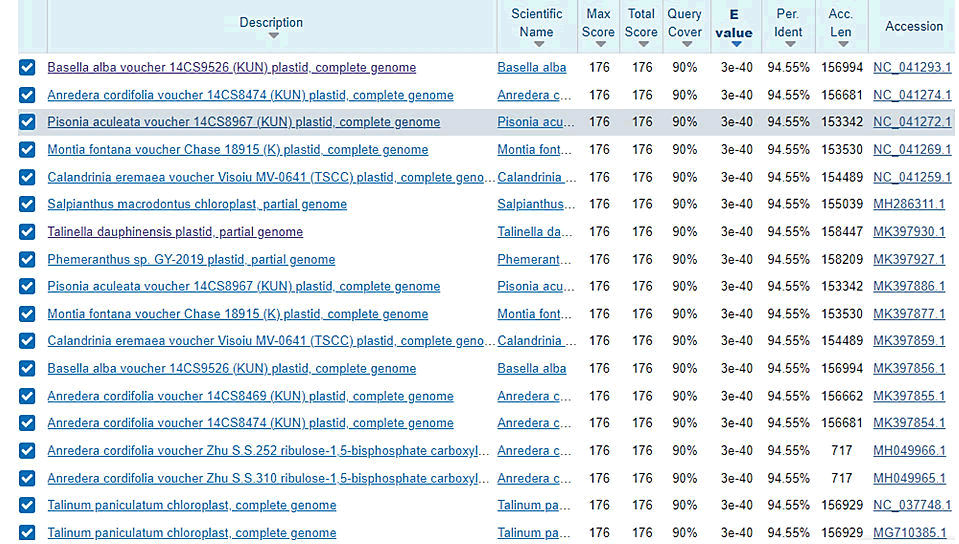
Figure 4: Blastn sequence search for 1b forward sequence indicating the outcome of similarities with their % identities.

Figure 5: Basellaalba voucherblastn search for forward sequence indicating high similarity.
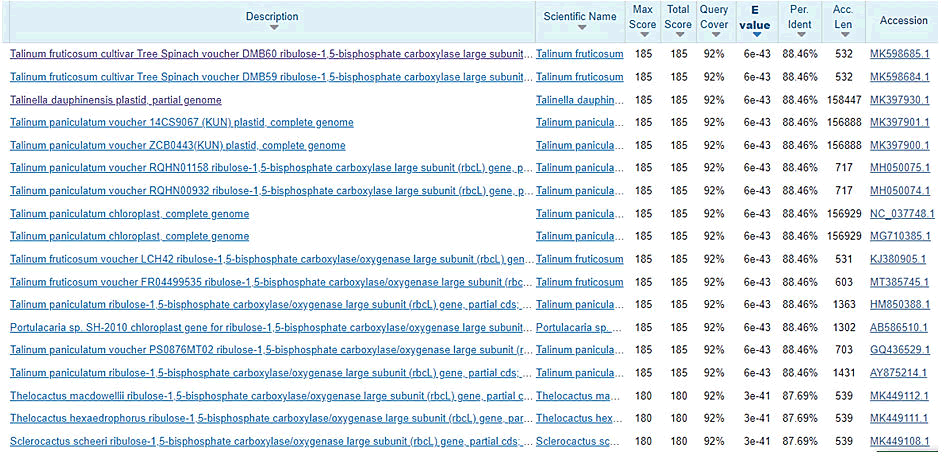
Figure 6: Blastn sequence search for 1b reverse sequence indicating the outcome of similarities with their % identities.
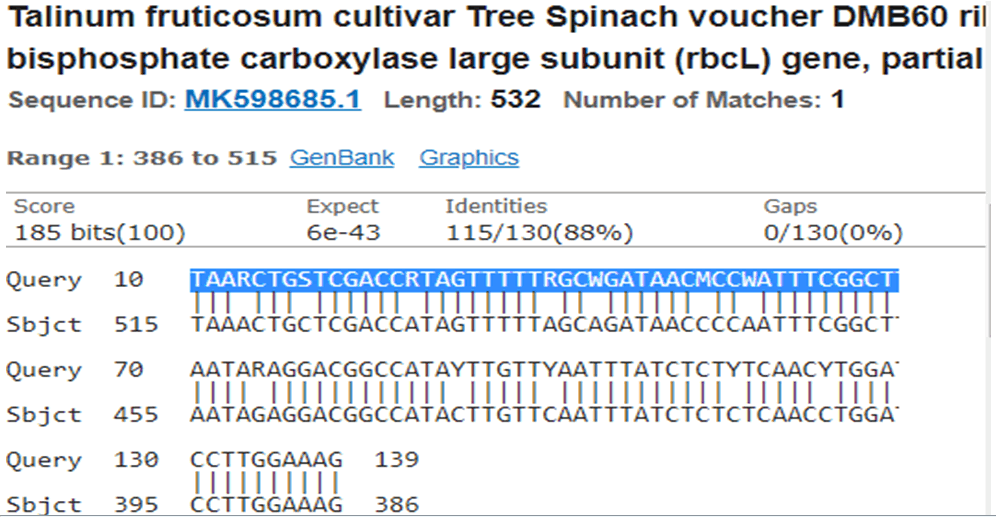
Figure 7: Talinum fruticosum cultivarblastn search for 1b reverse sequence indicating high similarity.
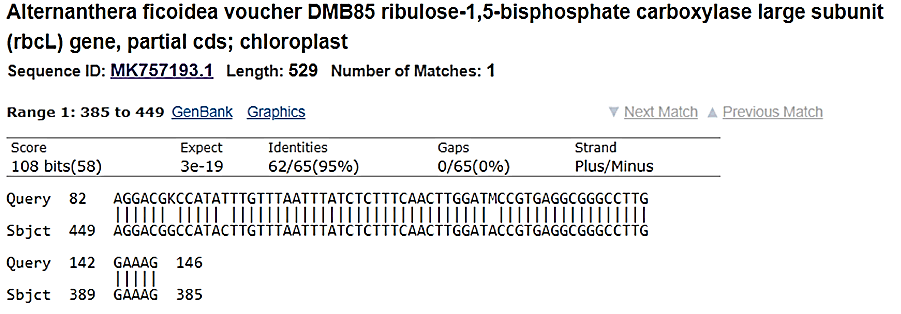
Figure 8: Alternanthraficoidea voucherblastn search w ith 1c reverse sequence indicating high similarity.
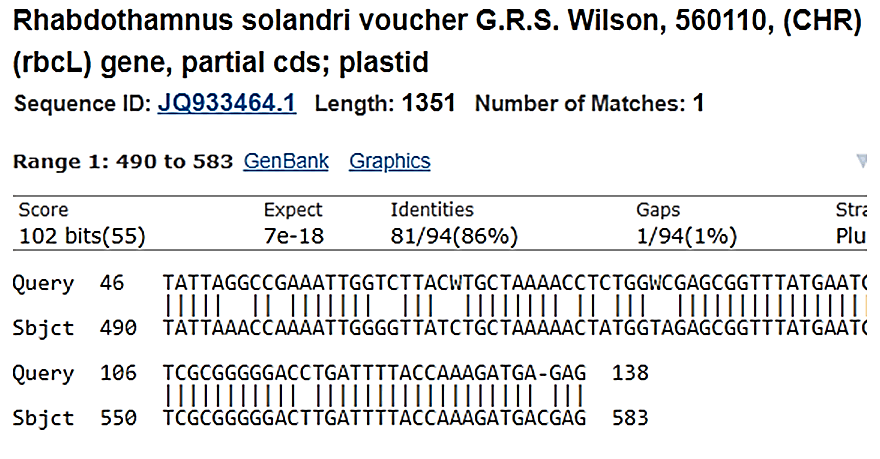
Figure 9: Rhabdthamnus solandri voucher search 1d reverse sequence indicating high similarity.
Morphological Description of Gnetum africanum
Morphological characters are shown in the degree of differences expressed in colors and shape of leaves, stems, flowers and pods in the land races and could be very useful as genetic markers. Differences in the floral structure, inflorescence types, peduncle, growth habit and style inserted or exerted could also serve similar purposes in the characterization of germplasm and in the studies of inheritance. Distinct characters among the different accessions were investigated using morphological descriptors according. The three different accessions of Gnetum africanum were analyzed for morphological variation based on the phenotypic expressions of the plant samples collected.
The samples collected from different geographical areas within Akwa Ibom State were completely different by their leaves shapes, size and colour. The Etinan sample appeared healthy with big round vines, leaves which were succulent in nature, while Ekim sample had smaller long leaves which were dark green in colour and Ikot Akpaden sample were a light green with much smaller round leaves was observed in the sample which make the sample look much tender in nature. These distinct features observed in the plants could be attributable to varying environmental factors and soil conditions, excessive sunlight and water availability in the locations from where they were raise and grown. For a long time, morphological information has been the basis for characterization of rich plants diversity. Nevertheless, there remains considerable concern about the precise role of morphology in characterization as they are inadequate for definite identification of plants up to species level, due partly to fluctuation in environmental and soils condition.
Furthermore, these morphological descriptors are often restricted as the characters may not be apparent at all stages of the plant development as growth routine may vary. Thus, this calls for the use of additional descriptors involving in depth analysis at the molecular level as essential to identifying and authenticating the rich diversity of plants.
Molecular Characterization
Molecular descriptors provide valuable information on genetic diversity in view of their ability to decipher variation existing at the DNA level. Identification of any plant up to species level is of fundamental importance in diversity studies. This showed that for the assessment of plants diversity, it is essential that individual plant be precisely classified to species level. Furthermore, to establish effective management of plant genetic diversity for sustenance and maintenance of plant Germplasm, it is imperative to regard genetic variations at both interspecific and intraspecific levels as ecological asset and richness or affluence. The genetic compositions of plant types are always distinct. In view of this, their discrimination from their wild relatives is important in the development of appropriate conservation study. Depending on the stage of our development genetic diversity may be considered at different organizational levels of the gene pool, individual or at population level, locus and DNA level/ based sequence.
DNA Quality Analysis on Electrophoresis
The isolation of plant DNA is the first step for any type of molecular analysis. In the present study, crisp DNA bands without any trim were observed in agarose gel for all the 3 samples, shown as a clear band at 390 bp. The results shown in the present study demonstrates that the 3 accessions using the rbcl gene as primer was successfully amplified and that was seen or shown as a clear band at 330 bp, this was not withstanding the differences in the geographical distance associated with each accession. Thus, the intra specific diversity could be due to individual species response to the changes in environmental conditions from where they were collected. This realization calls for further analysis to investigate the genetic conformity of the different accessions.
DNA Quantity Analysis on Nano Drop Spectrophotometer
In this study, DNA template from the different accessions of Gnetum africanum was assessed on a nano drop spectrophotometer for their concentrations. The results obtained were a clear indication of the genetic composition existing among the species. This high genetic content shown across the three species of Gnetum africanum could be attributable to the geographical distributions of its landraces and evolutionary history of the liana. Although the samples were all picked from one ecological zone of the tropical rainforest they represent accessions collected from different regions with different agro-ecologies within the country, Nigeria.
DNA Barcoding and DNA Sequencing of the Amplified Product
In this study, DNA barcoding and sequencing was carried out and rbcL barcode was used to discriminate the three accession of Gnetum africanum. Most authors report the availability of DNA sequence information for the complete coding region of plant DNA. This also informs us for embarking on barcoding study for the different accessions of Gnetum africanum to ascertain their genetic conformity. The results of the sequencing of the amplicons and blast analysis showed no variation among the different accessions or samples. This similarity in genotypic data existing among the three accession of Gnetum africanum at species level has been reported earlier with Talinum triangulare. The rbcL amplified product of Gnetum africanum (330bp) showed up to 95% identity with Basellaalba voucher NC 041293.1 with 90% query coverage for forward sequence and 88% similarity with Talinum fruticosum cultivar MK 598685.1 with 92% query coverage.
The study was aimed at characterizing Gnetum africanum land races to identify desirable characters in the accession that could be used for the development of high yielding hybrids for domestication In this study, DNA barcoding and sequencing was carried out and rbcL barcode was used to discriminate the three accession of Gnetum africanum. Most authors report the availability of DNA sequence information for the complete coding region of plant DNA. This also informs us for embarking on barcoding study for the different accessions of Gnetum africanum to ascertain their genetic conformity. The results of the sequencing of the amplicons and blast analysis showed no variation among the different accessions or samples.
The authors declare that they have no conflict of interest.
Citation: Nya E, Owoh L, Udofia O, Udosen I, Ogidi EG, et al. (2022) Morphological and Molecular Characterization of Gnetum africanum (Welw) Germplasm Using DNA Barcoding Method. Eur Exp Bio. 12:82.
Copyright: © 2022 Nya E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.