Keywords
TREX-2; EMSA; RNP particle
Introduction
Origin recognition complex (Orc) was first discovered in yeast
cells where its function is to recognize the replication initiation
site and to guide the replication complex to the replication origins
[1]. Later, homologs of the yeast Orc proteins have been found
in all eukaryotic organisms [2-4]. It has also been demonstrated
that unlike yeast Orc, the Orc proteins in higher eukaryotes have
additional functions distinct from their role in replication initiation
[5-8]. For instance, in our previous work we have demonstrated
that in Drosophila, Orc interacts with the TREX-2/THSC/AMEX
complex involved in the mRNA transport from the nucleus to the
cytoplasm [9].
The latter complex was originally detected in yeast and includes
Sac3 protein, two molecules of Sus1 protein, as well as Cdc31 and
Sem1p proteins [10]. Orthologs of the TREX-2 complex have been
found in higher eukaryotes [11,12]. Sac3 (Xmas-2 in Drosophila)
within the complex, provides a platform for the interaction with
other subunits, as well as with the complex-associated proteins.
The N-terminal part of Sac3 is responsible for the mRNA binding
and interacts with other proteins in the complex which are
involved in the interactions with mRNA, namely, Thp1 and Sem1
[13-15]. The C-terminal part of Sac3 interacts with the two copies
of Sus1 and with Cdc31, this sub-complex being responsible for
the interactions with the nuclear pore [15,16].
In our laboratory, we have described the Drosophila TREX-2
complex and its individual subunits, Xmas-2 and ENY2. We have
demonstrated that Xmas-2 and ENY2, the orthologs of the yeast
Sac3 and Sus1 proteins, respectively, interact with each other and
account for the mRNA export to the cytoplasm [16]. We have also
shown that ENY2, as a part of the TREX-2 complex, is associated
with the nuclear pore [16]. It was demonstrated further that Orc
interacts with the TREX-2 complex. Orc subunits also interact
with the mRNP complex and participate in mRNP export to the
cytoplasm. Orc3 subunit is the strongest interacting subunit out
of all Orc subunits, in the Orc association with the TREX-2 complex
and mRNP particle [9]. In particular, this subunit most efficiently
interacts with the Xmas-2 and ENY2 components of TREX-2 [9].
In the current work, we studied interactions of Orc3 with the Xmas-
2 and ENY2 components of the TREX-2 complex. We found that
Orc3 was associated with Xmas-2 through two molecules of ENY2.
The EMSA with the labeled fragments of the ras2 gene mRNA,
demonstrated that Drosophila Orc3 directly interacts with mRNA
with a low specificity. Our data suggest that Orc3 is most likely
responsible for assembling the Orc, TREX-2, and mRNP complexes
together.
Methods
Cloning of protein expression constructs
Coding sequences for the full-size Xmas-2 and ENY2 proteins with
the C-terminally tagged HA epitope and His epitope, and coding gene
regions, corresponding to the Orc3 N-terminal domain (1-333 aa),
Orc3 C-terminal domain (334-721 aa), and full-size Orc3 N-terminally
fused with three FLAG epitopes and GST epitope were cloned into
pAc5.1/V5-His vector (Invitrogen). Vector constructs containing the
coding regions of the Orc3 gene, corresponding to the protein’s
N-terminal and C-terminal domains fused with GST epitope, were
generated.
Transfection, expression, and purification of
recombinant proteins
Transfection of Scheider-2 cells (S2) was performed using the
Effectene Transfection Reagent as described in [17]. Expressions of
the recombinant GST-tagged proteins were performed in E. coli BL21
cells at 20°C for 24 h. Cell were collected by centrifugation and stored
at −70°C. Recombinant proteins with the GST epitope tag were
purified from the cell lysates by binding to glutathione Sepharose and
subsequently eluted by displacement with glutathione, according to
the manufacturer’s recommendations (GE Healthcare).
GST pull-down assay
GST-fused protein immobilized on the glutathione Sepharose 4
fast flow resin (GE Healthcare) was washed extensively with LBST-
100 buffer (50 mM Tris [pH-8.0], 100 mM NaCl, 5 mM MgCl2, 20
% glycerol, 0.1% Triton X-100, and 0.5 mM DTT). His-E(y)2 or His-
Xmas-2 proteins were added to the resin, and the interaction
assay was carried out in a final volume of 600 mL. After incubation,
beads were washed five times with increasing NaCl concentrations
(LBST-100, LBST-300, and LBST-500) [18]. Western blotting and
immunoprecipitation techniques were carried out using standard
protocols.
Drosophila cell culture extracts
Drosophila S2 cells were maintained at 25°C in the Schneider’s
insect medium (Sigma) containing 10% fetal bovine serum
(HyClone, United States). To extract proteins, S2 cells were
lysed. Cells were centrifuged at 2000 rpm at +4°C for 5 min and
resuspended in 1 ml of 1x PBS with 25x PIC (Protease Inhibitor
Cocktail, Roche). Cell were then centrifuged once more at 2000
rpm for 5 min at 4°C and resuspended in LB buffer, containing 10
mM Hepes pH-7.0, 0.4 M NaCl, 5 mM MgCl2, 0.5% NP-40, 25x PIC,
1 mM DTT, and 0.3 μL of DNAse I, at the ratio of 1/10. Cells were
incubated in LB buffer for 20 min on ice and the lysate was further
centrifuged at 13000 rpm for 15 min at 4°C.
Drosophila embryonic nuclear extract
The nuclear material was extracted from 0- to 12-h old Drosophila
embryos with 0.42 M ammonium sulfate solution, as described [19].
Immunoprecipitations
Antibodies were bound to protein A Sepharose (Sigma) following
the manufacturer’s recommendations. 100 μL of the protein extract in IP buffer (10 мМ HEPES, рН 7.9, 5 мМ MgCl2, 0.1% NP-
40, 0.15 М NaCl, 25x PIC, 1 mM DTT, RNase (Stratagene, 10 U/mL)
0.5 ng/μL of DNAse I) were incubated with 10 μL of Sepharose
overnight at 4°C. Then, Sepharose was sedimented, supernatant
was discarded, and the precipitate was washed three times with
100 μL of IP 500 buffer (10 мМ HEPES, рН 7.9, 5 mM MgCl2, 10%
glycerol, 500 mM KCl, and 0.1% NP-40) for 10 min each. The
equal volume of the electrophoresis loading buffer was further
added to the precipitate, the obtained suspension was boiled,
centrifuged, and supernatant was applied to the polyacrylamide
gel for Western blotting.
Antibodies
Polyclonal antibodies against Orc3 were described previously [9].
Anti-GST polyclonal antibodies were purchased from Santa Cruz
Biotechnologies. Antibodies against Xmas-2 and ENY2 have been
described previously [16].
Synthesis of the radiolabeled RNA fragments
DNA templates for the α-32P-RNA synthesis of the ras2 mRNA
fragments were prepared from pSK-ras plasmids by linearization
of the cloned sequence from the 3’-end for the sense fragments.
Radiolabeled RNAs were prepared using the RNA Labeling Mix and
T7 and T3 polymerases (Roche Diagnostics), treated with RNasefree
DNase I, and purified using the RNeasy Kit (Qiagen). Each
radiolabeled RNA was analyzed by agarose gel electrophoresis
and quantified by UV-spectrometry..
Electrophoretic mobility shift assay (EMSA)
Purified recombinant proteins corresponding to the N-terminal
and C-terminal parts of Orc3 were incubated with the radiolabeled
RNA fragments in 20 μL of binding buffer, containing 25 mM Tris-
HCl, pH 8.0, 100 mM NaCl, 0.5% Triton X-100, 5% glycerol, 1 mM
EDTA, 1 mM DTT, 25x PIC, 40x RiboLock (ThermoScientific) for 1
h at 4°C. After binding was completed, PAGE loading buffer was
added to the samples, and samples were applied to the 5% native
polyacrylamide gel. Electrophoretic separation was carried out in
0.5x TBE buffer at 150 V for 80 min at 4°C. Radioactive signal was
detected with the aid of the Cyclone StoragePhosphor Screen
device. The signal in each line was quantitated using ImageJ
program. Direct titration reactions were plotted as fraction of
bound RNA versus the protein concentration and KD for each
fragments were determined according to [20,21].
Results
Analysis of the Orc3 interaction with the TREX-2
complex subunits ENY2 and Xmas-2
Earlier, in the experiments with studying protein co-precipitations
from the nuclear extracts, we have shown that Orc3 readily coprecipitates
with both the Xmas-2 and the ENY2 subunits of
the TREX-2 complex [9]. Xmas-2 (170 KDa) is the main subunit
of the TREX-2 complex and it interacts via its C-end with two
molecules of ENY2 (11 KDa). The direct interaction of Drosophila
Xmas-2 and ENY2 was demonstrated [16]. In the current work,
we have studied the interactions of the Orc3 protein with these
two subunits in more detail. The domain organization of Orc3 is highly conservative. It is tentatively subdivided into two parts, the
N-terminal part which corresponds to an AAA+-like domain and
contains motifs mediating protein interactions with nucleic acids,
and the C-terminal part [22]. Earlier, some authors investigated
interactions of Orc3 with other components of the Orc complex
[22,23]. They have demonstrated that the N-terminal part of
Orc3, representing the AAA+-like domain, interacts only with
the C-terminal domain of Orc2 and with no other subunits of
the complex [22]. In the C-terminal region of Orc3, there was
identified an “Insertion” domain mediating an interaction with
the Orc6 protein [23], as well as a WH domain involved in the interactions
with Orc4, Orc5, and some other proteins (Figure 1A) [22].
To study interactions of Orc3 with the components of the TREX-
2 complex, we split the Orc3 protein into two protein moieties,
one of which (Orc3N) corresponded to the AAA+ domain and the
other (Orc3C) contained the “Insertion” and the WD domains.
This split was performed in order to determine which region of
Orc3 contains the domain responsible for its interaction with the
Xmas-2 and ENY2 subunits of the TREX-2 complex. We generated
several expression constructs for the proteins representing the
Orc3 N-terminal domain, or the Orc3 C-terminal domain, each
fused with the FLAG epitope (Orc3N and Orc3C) in the Drosophila
S2 cells. In these series of experiments, we also used expression
constructs for Xmas-2 and ENY2 proteins fused with the HA
epitope and the Orc3 protein fused with the FLAG epitope. To
study interactions between Orc3 and Xmas-2, Drosophila S2 cells
were double transfected with the following pairs of expression
constructs: Orc3N-FLAG and Xmas-2-HA, Orc3C-FLAG and Xmas-
2-HA, and Orc3-FLAG and Xmas-2-HA.
Protein co-immunoprecipitations from the S2 cell lysates
(Figure 1B) did not reveal any interactions between Orc3C
and Xmas-2, and between the full-size Orc3 and Xmas-2. No
interactions were observed between the N-terminal part of Orc3
and the Xmas-2 protein. Therefore, most likely, there are no
significant interactions between these two proteins, under these
conditions.
To study interactions between Orc3 and ENY2, S2 cell were
double transfected with pairs of genetic constructs for the
expression of Orc3N-FLAG and ENY2-HA, Orc3C-FLAG and ENY2-
HA, as well as Orc3-FLAG and ENY2-HA. Co-immunoprecipitations
with antibodies against the HA epitope and immunostainings
of Western-blots with antibodies against FLAG (Figure 1C),
demonstrated that ENY2 interacted with both the N-terminal
domain of Orc3 and its C-terminal region. However, it revealed a
higher affinity for the C-terminal domain.
This indicates that each of the two Orc3 regions interacts
with a single ENY2 molecule. Therefore, since we detected
no interactions between Orc3 and Xmas-2, it is likely that one
molecule of Orc3 interacts with two molecules of ENY2. Our
data demonstrate that the interaction between Xmas-2 and Orc3
proteins, observed in the co-immunoprecipitation experiments
of endogenous proteins from the nuclear extract, is not a direct
one but is mediated by the ENY2 protein.
In line with this finding, antibodies against Orc3 efficiently
co-precipitated ENY2 from the nuclear extract of Drosophila
embryos, and vice versa (Figure 1D). In addition, we investigated
whether interaction of Orc3 and ENY2 was direct in the GST pulldown
assay (Figure 1E). The bacterially expressed GST-tagged
Orc3 was immobilized on glutathione Sepharose. The His-tagged
ENY2 was also expressed in bacteria, purified, and incubated with
the glutathione Sepharose bound Orc3. Our data demonstrate
that ENY2 strongly interacts with Orc3. In our experiments we did
not observe a direct interaction of Orc3 with Xmas-2.
Xmas-2, the largest subunit of the Drosophila TREX-2 complex,
forms the scaffold of the complex, while two ENY2 molecules bind
with its C-terminal part (Figure 1F). Orc3, presumably, interacts
with the TREX-2 complex via binding to some motifs located in
its N- and C-terminal parts, with the two ENY2 molecules present
within the complex (Figure 1F).

Figure 1: Analysis of Orc3 interactions with the ENY2 and Xmas-2 proteins. A: Schematic representation of the Drosophila Orc3 protein
domain organization. Orc3N -- N-terminal domain of Orc3 (350 aa); Orc3C -- C-terminal domain of Orc3 (371 aa); AAA+-like --
Orc3 domain responsible for binding with nucleic acids and the Orc2 protein; Insertion -- Orc3 domain responsible for binding
with the Orc6 protein; WH -- Orc3 domain necessary for the interaction with different proteins [22, 23]. B and C: Western blot
analysis of Orc3 interactions with the ENY2 and Xmas-2 proteins. B, Western blot analysis of the interactions between the
Xmas-2 and Orc3N, Xmas-2 and Orc3C, and Xmas-2 and full-size Orc3 proteins over-expressed pairwise in the Drosophila S2
cells. C, Western blot analysis of the interactions between the ENY2 and Orc3N, ENY2 and Orc3C, and ENY2 and full-size Orc3
proteins over-expressed pairwise in the Drosophila S2 cells. D: The co-immunoprecipitation of Orc3 and ENY2 from the nuclear
extract of Drosophila embryos and vice versa. E: The interaction of Orc3 and ENY2 or Xmas-2 in the GST pull-down assay. The
bacterially expressed GST-tagged Orc3 was immobilized on glutathione Sepharose. The His-tagged ENY2 or His-tagged Xmas-2
were expressed in bacteria, purified, and mixed with the glutathione Sepharose4 bound Orc3 in the incubation buffer. 15 μL of
this mixture was loaded onto the gel as Input. After the incubation glutathione Sepharose4 resin was washed and GST-bound
proteins were eluted, 15 μL of eluates were loaded onto the gel (Eluate). Western blot was developed with antibodies against
GST, ENY2, or Xmas-2. F: The schematic representation of presumable interactions between Orc3, ENY2, and Xmas-2.
Orc3 protein directly binds to the mRNA of the
ras2 gene
We have already demonstrated earlier that Orc subunits interact
with the mRNP of the ras2 gene [9], with the Orc3 protein
demonstrating the highest affinity to ras2 mRNP comparatively to
all the other subunits. In the current work, we aimed to elucidate
whether the Drosophila Orc3 protein directly interacts with
mRNA. And if such an interaction takes place, which part of the
protein, the N-terminal region corresponding to the AAA+-like
domain, or the C-terminal region, is necessary for this interaction.
Lastly, we wanted to determine which region of the ras2 mRNA
interacts with Orc3.
In this series of experiments, we used the polyacrylamide gel
electrophoretic mobility shift assay (EMSA). The nucleotide
sequences corresponding to the N-terminal and C-terminal
regions of Orc3 were cloned into the pGEX vector in frame with
the GST epitope. The target proteins were expressed in a bacterial
system and were purified using glutathione Sepharose. The ras2
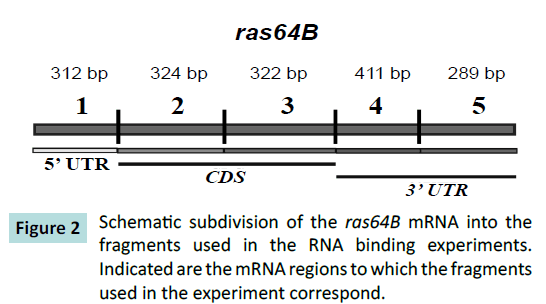
mRNA sequences were subdivided into five fragments (Figure 2).
To synthesize sufficient amounts of the target mRNA fragments
the corresponding DNA sequences were cloned into the pSK
vector containing T3 and T7 promoters. For each DNA fragment,
a radioactively-labeled RNA was synthesized using T3 or T7 RNA
polymerase and α-32P UTP. The obtained radiolabeled RNAs were
incubated in the binding buffer with the proteins corresponding
either to the N-terminal or to the C-terminal domain of the Orc3
protein.
Figure 2: Schematic subdivision of the ras64B mRNA into the
fragments used in the RNA binding experiments.
Indicated are the mRNA regions to which the fragments
used in the experiment correspond.
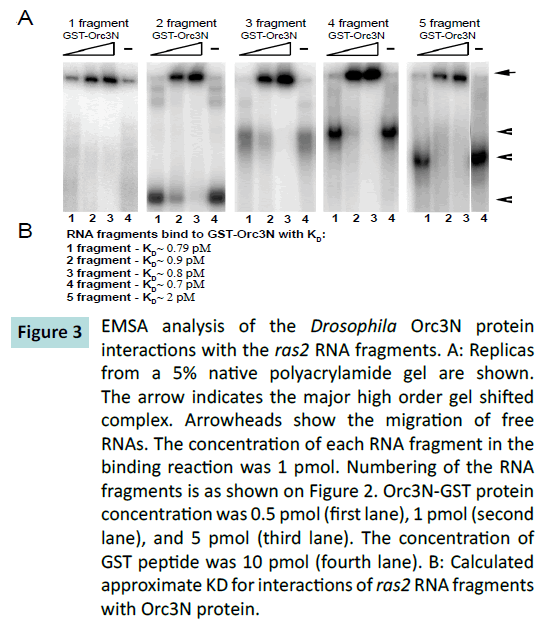
We observed that the Orc3 N-terminal region corresponding
to the AAA+-like domain, interacted with all the fragments
of the particular ras2 mRNA (Figure 3). At the same time, GST
peptide did not interact with the RNA fragments by itself (lane
4), thus validating specificity of our experiments. Therefore,
our experiments have demonstrated that Orc3 directly binds to
mRNA.
Figure 3: EMSA analysis of the Drosophila Orc3N protein
interactions with the ras2 RNA fragments. A: Replicas
from a 5% native polyacrylamide gel are shown.
The arrow indicates the major high order gel shifted
complex. Arrowheads show the migration of free
RNAs. The concentration of each RNA fragment in the
binding reaction was 1 pmol. Numbering of the RNA
fragments is as shown on Figure 2. Orc3N-GST protein
concentration was 0.5 pmol (first lane), 1 pmol (second
lane), and 5 pmol (third lane). The concentration of
GST peptide was 10 pmol (fourth lane). B: Calculated
approximate KD for interactions of ras2 RNA fragments
with Orc3N protein.
The N-terminal region of Orc3 similarly interacted with all the
RNA fragments, suggesting that there is no strong preference
for interaction with a certain ras2 mRNA region. However, this
Orc3 protein region showed the highest affinity for the first and
the fourth ras2 gene fragments (Figure 3B), comparatively to the other fragments. This may indicate that there is still some level of
specificity in the interaction with these RNA fragments (Figure 3).
Fragment 1 comprises the 5’-noncoding region of the gene, while
fragment 4 corresponds to the proximal 3’-noncoding region.
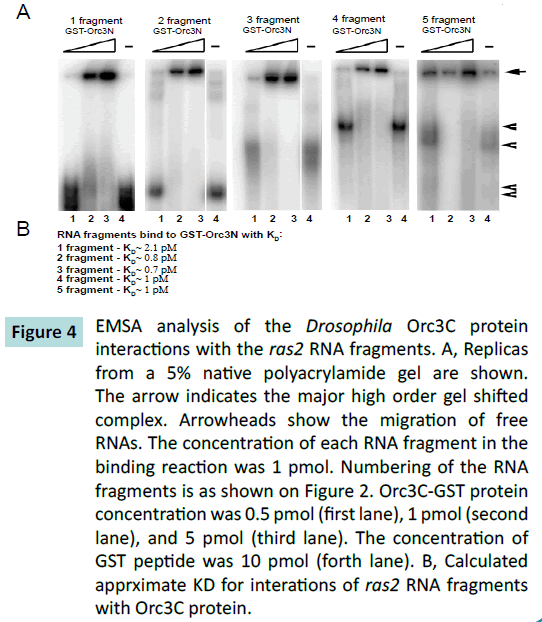
The C-terminal domain of Orc3 also interacted with all the ras2
mRNA fragments. The strongest interaction appeared to be with
the third fragment, which corresponds to the distal part of the
coding region of the gene (Figures 4A and 4B) .
Figure 4: EMSA analysis of the Drosophila Orc3C protein
interactions with the ras2 RNA fragments. A, Replicas
from a 5% native polyacrylamide gel are shown.
The arrow indicates the major high order gel shifted
complex. Arrowheads show the migration of free
RNAs. The concentration of each RNA fragment in the
binding reaction was 1 pmol. Numbering of the RNA
fragments is as shown on Figure 2. Orc3C-GST protein
concentration was 0.5 pmol (first lane), 1 pmol (second
lane), and 5 pmol (third lane). The concentration of
GST peptide was 10 pmol (forth lane). B, Calculated
apprximate KD for interations of ras2 RNA fragments
with Orc3C protein.
To summarize, both of the Orc3 regions were able to bind to the
ras2 RNA. Our data suggest that Orc3 possesses several RNAbinding
domains localized in both parts of the protein..
Discussion
Here, we studied interactions of the Orc3 protein, a subunit of
Orc complex, with the Xmas-2 and ENY2 subunits of the TREX-2
complex and have demonstrated that Orc3 directly interacts with
ENY2. These interactions were carried out by both the N-terminal
domain and the C-terminal domain of Orc3.
Earlier, we have purified the Drosophila mRNA export complex,
TREX-2, and found that it is associated with the Orc complex
[9]. However, Orc2 was the only Orc subunit which did not copurify
with TREX-2. Orc2 interacts with the Orc complex via its
Orc3 subunit. As was demonstrated previously, human Orc3
N-terminal domain interacts with Orc2, while its C-terminal
domain is required for the interaction with Orc4 and Orc5 [22].
Here, we have found that the N-terminal domain of Orc3 could also interact with TREX-2. These data suggest that TREX-2 may
replace Orc2 when it interacts with Orc and this may explain why
Orc2 was not detected within the Orc-TREX-2 complex.
To study interactions of Orc3 with the Xmas-2 and ENY2 subunits
of the TREX-2, we over-expressed two proteins, one corresponding
to the N-terminal region of Orc3, and the other corresponding to
its C-terminal region. These proteins were co-expressed pairwise
with the ENY2 and Xmas-2 proteins in the Drosophila S2 cells.
Both fragments of the Orc3 protein, as well as the full size Orc3,
interacted with ENY2. We suppose that the interactions between
the three proteins, Xmas-2, ENY2, and Orc3, within the Orc-
TREX-2 complex, are mediated by the interactions between Orc3
and ENY2, with both of the Orc3 domains being involved. It is
well documented that TREX-2 complex contains two molecules
of ENY2 [13,14]. It is thought that one of these molecules is
involved in the interaction of TREX-2 with the nuclear pore
complex [10,13,14,24,25]. It seems possible that Orc3 binds with
both copies of the ENY2 protein and, if this were the case, it could
also be involved in the interaction with the nuclear pore complex.
However, this latter conjecture requires further study.
The Orc3 structure and function analysis revealed a domain
potentially responsible for the interaction with nucleic acids.
This domain encompasses two amino acid motifs capable of
binding ATP or GTP. This nucleic acid interaction domain is
localized in the N-terminal part of Orc3 and is called the AAA-like
domain [26,27]. Traditionally, Orc proteins are known for their
interaction with DNA and their initiation of the DNA replication
[28]. Nevertheless, recently it has emerged that these proteins
can also interact with RNA [29-33]. In our previous study using
RNA immunoprecipitation, we have demonstrated that ORC
proteins interact with the mRNP of several genes [9]. Here, we
assessed the interactions of the Orc3 N-terminal and C-terminal domains with different fragments of the ras2 mRNA. We found
that both of the Orc3 termini are able to interact with the RNA
fragments, suggesting that Orc3 has an RNA-binding ability. This
also indicated that in the interactions between Orc and the mRNP
particle, described by us previously [9], it is mRNA to which Orc3
binds directly, and not only the protein components of mRNP.
In the current work, we also studied which particular regions
of the ras2 mRNA interact with Orc3. We found that both Orc3
domains show no specificity in the interaction and bind to any of
the RNA fragments. Interestingly, the N-terminal protein domain
of Orc3 showed different affinity for RNA, than the C-terminal
region. The N-terminus had the highest affinity to the RNA
fragments 1 and 4, corresponding to the 5’-noncoding region and
to the 3’-proximal part of the non-coding region, respectively. At
the same time, the protein corresponding to the Orc3 C-terminal
domain bound with the highest affinity to the different parts of
the coding region of the ras2 mRNA.
We hypothesize that Orc3 protein contains another, yet
unidentified nucleic acid-binding domain which is located in its C-terminal part. Since the two truncated proteins show their
maximum affinity to different mRNA fragments, it could be
that the full-size native protein interacts with RNA by its both,
N-terminal and C-terminal, domains which bind different mRNA
regions to ensure a closer contact with the protein moiety of the
mRNA-containing complex.
Acknowledgment
The work was supported by the Russian Scientific Foundation,
grant no. 14-14-01059.
Conflict of Interest
The authors declare that they have no conflicts of interest with
the contents of this article.
Author Contributions
VVP carried out first part of experiments, DVK – carried out EMSA
experiments and wrote most of the paper. SGG conceived the
idea for the project and wrote the paper with DVK.
References
- Bell SP,Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333-374.
- Bell SP(2002) The origin recognition complex: from simple origins to complex functions. Genes Dev 16: 659-672.
- Gossen M,Pak DT,Hansen SK,Acharya JK,Botchan MR(1995) A Drosophila homolog of the yeast origin recognition complex. Science 270: 1674-1677.
- Vashee S,Simancek P,Challberg MD,Kelly TJ(2001) Assembly of the human origin recognition complex. J Biol Chem 276: 26666-26673.
- Carpenter PB,Mueller PR,Dunphy WG(1996) Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature 379: 357-360.
- Chesnokov IN,Chesnokova ON,Botchan M(2003) A cytokinetic function of Drosophila Orc6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci USA 100: 9150-9155.
- Romanowski P,Madine MA,Rowles A,Blow JJ,Laskey RA(1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol 6: 1416-1425.
- Rowles A,Chong JP,Brown L,Howell M,Evan GI, et al. (1996) Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell 87: 287-296.
- Kopytova D,Popova V,Kurshakova M,Shidlovskii Y,Nabirochkina E,et al. (2016) Orc interacts with THSC/TREX-2 and its subunits promote Nxf1 association with mRNP and mRNA export in Drosophila. Nucleic Acids Res.
- Rodriguez-Navarro S,Fischer T,Luo MJ,Antunez O,Brettschneider S,et al. (2004) Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75-86.
- Dimitrova L,Valkov E,Aibara S,Flemming D,Mclaughlin SH, et al. (2015) Structural characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2. Structure 23: 1246-1257.
- Pascual-Garcia P,Govind CK,Queralt E,Cuenca-Bono B,Llopis A, et al. (2008) Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev 22: 2811-2822.
- Ellisdon AM,Dimitrova L,Hurt E,Stewart M(2012) Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nat Struct Mol Biol 19: 328-336.
- Jani D,Lutz S,Marshall NJ,Fischer T,Kohler A,et al. (2009) Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell 33: 727-737.
- Jani D,Valkov E,Stewart M(2014) Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res 42: 6686-6697.
- Kurshakova MM,Krasnov AN,Kopytova DV,Shidlovskii YV,Nikolenko JV,et al. (2007) SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J 26: 4956-4965.
- Vorobyeva NE,Soshnikova NV,Nikolenko JV,Kuzmina JL,Nabirochkina EN,et al. (2009) Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc Natl Acad Sci USA 106: 11049-11054.
- Kurshakova M,Maksimenko O,Golovnin A,Pulina M,Georgieva S, et al. (2007) Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol Cell 27: 332-338.
- Georgieva S,Nabirochkina E,Dilworth FJ,Eickhoff H,Becker P, et al. (2001) The novel transcription factor e(y)2 interacts with TAF(II)40 and potentiates transcription activation on chromatin templates. Mol Cell Biol 21: 5223-5231.
- Goodrich JA,Kugel JF(2015) Studying the affinity, kinetic stability, and specificity of RNA/protein interactions: SINE ncRNA/Pol II complexes as a model system. Methods Mol Biol 1206: 165-178.
- Ryder SP,Recht MI,Williamson JR(2008) Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol Biol 488: 99-115.
- Dhar SK,Delmolino L,Dutta A(2001) Architecture of the human origin recognition complex. J Biol Chem 276: 29067-29071.
- Bleichert F,Balasov M,Chesnokov I,Nogales E,Botchan MR, et al. (2013) A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. Elife 2: e00882.
- Fischer T,Rodriguez-Navarro S,Pereira G,Racz A,Schiebel E, et al. (2004) Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol 6: 840-848.
- Jani D,Lutz S,Hurt E,Laskey RA,Stewart M,et al. (2012) Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res 40: 4562-4573.
- Liu J,Smith CL,Deryckere D,Deangelis K,Martin GS, et al. (2000) Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol Cell 6: 637-48.
- Speck C,Chen Z,Li H,Stillman B(2005) ATPase-dependent cooperative binding of Orc and Cdc6 to origin DNA. Nat Struct Mol Biol 12: 965-971.
- Chesnokov IN(2007) Multiple functions of the origin recognition complex. Int Rev Cytol256: 69-109.
- Deng Z,Norseen J,Wiedmer A,Riethman H,Lieberman PM(2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and Orc recruitment at telomeres. Mol Cell35: 403-413.
- Hoshina S,Yura K,Teranishi H,Kiyasu N,Tominaga A,et al. (2013) Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem 288: 30161-30171.
- Mohammad MM,Donti TR,Sebastian Yakisich J,Smith AG,Kapler GM(2007) Tetrahymena Orc contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J26: 5048-5060.
- Norseen J,Johnson FB,Lieberman PM(2009) Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol 83: 10336-10346.
- Norseen J,Thomae A,Sridharan V,Aiyar A,Schepers A, et al. (2008) RNA-dependent recruitment of the origin recognition complex. EMBO J 27: 3024-3035.