Research Article - (2024) Volume 9, Issue 3
Phenotypic Detection of erm Gene Encoding Methylase for Resistance to Macrolide, Lincosamide-Streptogramin-B (MSL-B) in Methicillin Sensitive and Methicillin Resistant Staph aureus Infections in a Tertiary Care Hospital of Nowshera
Hamzullah Khan1*,
Fazli Bari2,
Tayyaba Basharat3,
Mohammad Basharat Khan4,
Saadullah5 and
Shabbir Hussain6
1Department of Hematology, Charleston Medical College, Charleston, USA
2Department of Microbiology, Nowshera Medical College, Nowshera, Pakistan
3Department of Pharmacology, Nowshera Medical College, Nowshera, Pakistan
4Department of Chemical Pathology, Nowshera Medical College, Nowshera, Pakistan
5Department of Public Health, Sarhad University, Peshawar, Pakistan
6Department of Medicine, Nowshera Medical College, Nowshera, Pakistan
*Correspondence:
Hamzullah Khan, Department of Hematology, Charleston Medical College, Charleston,
USA,
Email:
Received: 29-Jul-2020, Manuscript No. IPPHR-24-5487;
Editor assigned: 03-Aug-2020, Pre QC No. IPPHR-24-5487 (PQ);
Reviewed: 17-Aug-2020, QC No. IPPHR-24-5487;
Revised: 01-Jul-2024, Manuscript No. IPPHR-24-5487 (R);
Published:
29-Jul-2024, DOI: 10.35841/2472-1646.9.02.14
Abstract
Objectives: Phenotypic detection of erm gene encoding enzyme in methicillin sensitive and methicillin
resistant Staphylococcus aureus isolates (sputum and pus samples) in a tertiary care hospital of
Nowshera and to find the sensitivity pattern to D+MSSA and D+MRSA.
Methods: This prospective cohort study was performed in the pathology laboratory of Qazi Hussain
Ahmed Medical Complex Nowshera from 1st March 2019 to 30th Sept 2019. A total of 186 S. aureus isolates were selected for routine antibiotic susceptibility testing. 52 samples showed D-test positive i.e., (induced clindamycine resistance). Relevant information were recorded on a predesigned
proforma prepared as per CLSI recommendation for data collection.
Results: Out of 186 patients Staph aureus isolates were selected for antibiotic susceptibility, 52
(27.95%) isolates showed inducible clindamycin resistance using D-test as per CLSI recommendations.
Out of total, 30 (57.7%) were females and 22 (42.3%) were males. The mean age with standard
deviation was 28.36 ± 3.8. The age range was from 15 years to 45 years of age. Mode of age was 23
years. In 45 (86.5%) cases D-test phenomenon was observed in MRSA while in 7 (13.5%) cases it was
also noted in Methicillin Sensitive Staph aureus (MSSA). 36 (69.23%) patients with phenotypically positive erm gene encoding methylase were referred from medical unit of QHAMC. The sensitivity
pattern to D+MRSA was; vancomycine 94.23%, lanezolid 94.23%, rifampicin 84.23%, fusidic acid
55.77%, doxycyclin 32.59%, levofloxacin 21.15%, gentamycine 13.46%, ciprofloxacin 13.46%.
It is pertinent to mention that the sensitivity to amoxicillin, cefradine, ceftriaxone was seen only in
cases with D+MSSA cases only (13.5%) cases. While no isolate sensitivity to co-trimaxazole.
The sensitivity to vancomycine and lanezolid was 100% in both D+MSSA and D+ MRSA but was not
reported in cases of D+MSSA to the clinicians to avoid its misuse of these precious drugs for simple D
+MSSA cases that can easily be treated with alternative b-lactam antibiotics.
Conclusion: D+MSSA cases where sensitive to b-lactam antibiotics and shall be treated with these
conventional antibiotics and precious drug like vancomycine and lanezolid should be kept reserved for
D+MRSA cases to avoid misuse of antibiotics and to reduce resistance. D+type of resistant infection
are now a challenge to the clinician to treat and is a public health threat that needs accumulative
response through advocacy, communication and social mobilization.
Keywords
Antibiotic resistance; MLS-antibiotics; Erm gene encoding methylase
Introduction
Staphylococcus aureus is a major notorious bacterial strand
causing nosocomial and community-acquired infections
around the globe. Furthermore an enhancing prevalence of
methicillin resistance among staphylococci is alarming. The
resistance to antibiotic used for the treatment of infection
caused by staphylococci is increasing with passage of time and
is challenging for the physician to treat. D-test is very easy and
cheaper test that involves disc diffusion (Kerby Baur) methods
and helps to study the Macrolide Lincosamide Streptogramin
resistance (MLSB) in Staphylococcus aureus. In this test, the
desk of macrolide i.e., erythromycin and a lincosamide extract i.e., clindamycin are placed adjacent to each other on Mueller
Hinton agar media and inoculated for interpretation.
Clindamycin and streptogramin are golden drugs used for
treating infections induced by Methicillin Resistant S. aureus (MRSA). Therefore resistance to this precious antibiotic in the
shape of D-test is annoying and challenging. It is pertinent to
mention that unlawful and irrational use of MLSB antibiotics
(macrolide-lincosamide-streptogramin B) is major contributing
factor to make the staphylococcal strains vigilant to acquire
resistance to MLSB antibiotics. Common most and generally
accepted mechanism of resistance to clindamycine is due to
mutation in the erm genes, that can be expressed two ways by
the klindamycin that is constitutively or inducibly. In both case
MLS-B strains are involved and in vitro it appears
erythromycin-resistant and clindamycin sensitive if the desks
of both are not placed adjacently. But the therapy with
clindamycine would result in failure. We conducted the study
with aim to find out frequency of inducible clindamycin
resistance in S. aureus infections from our geographical
location using D-test phenomenon.
Materials and Methods
This prospective cohort study was performed in the pathology
department of Qazi Hussain Ahmed Medical Complex
Nowshera from 1st Mar 2019 to 30th September 2019.
A total of 186 pus and sputum samples were received for
culture and sensitivity out of which 52 cases reported to be
phenotypically positive for erm-gene encoding methylase
(27.95%). The inclusion criteria were all cases irrespective of
age and gender received in the laboratory. Exclusion criteria
were samples received in the laboratory 24 hour after
collection, patient already on the antibiotic therapy and
improperly collected sputum and pus samples [1].
The samples were received in the pathology section from the
respective unit under observance of strict aseptic technique
after education of patients on pus and sputum sample
collection. Media were prepared as per CLSI (Clinical and
Laboratory Standard Institutes). All samples were inoculated
on selective medium MSA (Manitol Salt Agar). Then the
specimens were incubated under ambient air 35 ± 2 C for
18-20 hours. In case growth is obtained on MSA then further
inoculated on Mueller Hinton agar for sensitivity to antibiotics
as per CLSI recommendatios. The antibiotic desks used were;
VA-Vancomycine, LZD-Lanezolid, RD-Rifampicin, Fd-Fusidic
acid, Dox-Doxycyclin, Lev-Levofloxacin 21.15%, CNGentamycine,
Cip-ciprofloxacin, SXZ (Cotrimaxazole) and Foxcefoxetin.
Desk of erythromycin and klindamycine were placed at a
distance of 20 mm center to center. Phenotypically MSL
resistance was confirmed as Inhibition of zone of clindamycin
towards erythromycin as a strainght line, resembling the
alphabet “D” and was considered to be positive for D-test
phenomenon (Figure 1). Any haziness in the zone of inhibition
of clindamycin is also phenotypically representative of
resistance.
Plates were incubated for 18-20 hours and then zone of
inhibition were calculated with caliper, including the size of the
desk [2]. Zones were compared with CLSI recommendations for
sensitivity to be reported as sensitive, intermediate and
resistant. Finally the data obtained from the culture and
sensitivity was entered in a SPSS version 16 for descriptive and
correlation analysis of different parameters.
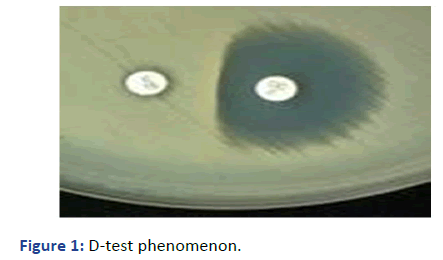
Figure 1: D-test phenomenon.
Results
A total of 186 samples were studied, 52 (27.9%) patients
showed D-test phenomenon, that were positive for erm-gene
encoding enzyme [3]. Out of those 30 (57.7%) were
females and 22 (42.3%) were males (Table 1).
| Variables |
Frequency |
Percent |
Valid percent |
Cumulative percent |
| Valid |
Female |
30 |
57.7 |
57.7 |
57.7 |
| Male |
22 |
42.3 |
42.3 |
100 |
| Total |
52 |
100 |
100 |
|
Table 1: Gender statistics.
The mean age with standard deviation was 28.36 ± 3.87. The
age range was from 15 years to 45 years of age [4].
Mode of age was 23 years (Table 2).
| N |
Valid |
52 |
| Missing |
0 |
| Mean |
28.3654 |
| Median |
25 |
| Mode |
23 |
| Std. deviation |
3.87145 |
| Range |
30 |
| Minimum |
15 |
| Maximum |
45 |
Table 2: Age statistics.
In 45 (86.5%) cases D-test phenomenon was observed in
MRSA while in 7 (13.5%) cases it was also noted in methicillin sensitive Staph aureus (Table 3).
| Variables |
Frequency |
Percent |
Valid percent |
| Valid |
MSSA D-positve |
7 |
13.5 |
13.5 |
| MRSA D-positve |
45 |
86.5 |
86.5 |
| Total |
52 |
100 |
100 |
Table 3: C/s report of phenotypic detection of erm gene encoding methylas in MSSA and MRSA.
36 (69.23%) patients with D-test positive were referred
from medical unit of QHAMC [5].
The rest of the patients were referred for culture
from surgical, pediatric and emergency units (Table 4).
| Unit |
Gender |
Total |
| Female |
Male |
| FMW |
CS report |
SA D-P positve |
2 |
|
2 |
| MRSA D-positve |
14 |
|
14 |
| Total |
16 |
|
16 |
| FSW |
CS report |
MRSA D-positve |
11 |
|
11 |
| Total |
11 |
|
11 |
| GYNE |
CS report |
MRSA D-positve |
2 |
|
2 |
| Total |
2 |
|
2 |
| MMW |
CS report |
SA D-positve |
0 |
4 |
4 |
| MRSA D-positve |
0 |
16 |
16 |
| Total |
0 |
20 |
20 |
| MSW |
CS report |
SA D-positve |
|
1 |
1 |
| MRSA D-positve |
|
2 |
2 |
| Total |
|
3 |
3 |
| Total |
CS report |
SA D-positve |
2 |
5 |
7 |
| MRSA D-positve |
28 |
17 |
45 |
| Total |
30 |
22 |
52 |
Table 4: Culture and sensitivity report sex unit cross tabulation.
The sensitivity pattern to D+MRSA was; vancomycine 94.23%,
lanezolid 94.23%, rifampicin 84.23%, fusidic acid 55.77%,
doxycyclin 32.59%, levofloxacin 21.15%, gentamycine 13.46%,
ciprofloxacin 13.46% [6].
It is pertinent to mention that the sensitivity to
betalactam antibiotics was seen only in cases with D
+MSSA (11.5%) cases [7]. while no sensitivity in any
both the groups seen for co-trimaxazole (Table 5).
| Antibiotics |
Sensitivity to D+MSSA positive cultures |
Sensitivity to D+MRSA Positive Cultures |
Total sensitivity |
Total cases |
Percentage |
| Vancomycine |
6 |
43 |
49 |
52 |
94.23 |
| Lanezolid |
5 |
44 |
49 |
52 |
94.23 |
| Rifampicin |
6 |
38 |
44 |
52 |
84.62 |
| Fusidic acid |
1 |
28 |
29 |
52 |
55.77 |
| Doxycycline |
3 |
14 |
17 |
52 |
32.69 |
| Levofloxacine |
2 |
9 |
11 |
52 |
21.15 |
| Gentamycin |
1 |
6 |
7 |
52 |
13.46 |
| Ciprofloxacin |
1 |
6 |
7 |
52 |
13.46 |
| Cefoxetin(b-lactam antibiotics) |
4 |
2 |
6 |
52 |
11.54 |
| Cotriamaxazole |
0 |
0 |
0 |
52 |
0 |
Table 5: Sensitivity pattern of D-test positive MSSA and MRSA.
The sensitivity to vancomycine and lanezolid was 100% in
both D+MSSA and D+MRSA but was not reported in cases of D
+MSSA to the clinicians to avoid its misuse of these precious
drugs for simple D+MSSA cases that can easily be treated with
betalactam antibiotics [8].
Discussion
The resistant strains of Staphylococcus aureus of emerged
soon after the initiation of antibiotics in early 1940s.
Macrolides, Lincosamides and Streptogramins (MLS) genes
are notorious for development of resistance to these
antibiotics in MSSA and MRSA infections. Resistance to
clindamycin induced by microclide mediated through a
methylase enzyme that alters the common ribosomal binding
site for macrolides, clindamycin and the group B
streptogrammins (quinupristin) makes Staph aureus more
resistant to another strand of antibiotic.
This emerging combined resistance of Staph aureus to
Macrolides, Lincosamides and Streptogramin-B (MLS-B) limits
the use of these drugs in infections caused by these bacteria.
In present study frequency of MLS-B phenotype in staph infections was 52/186 (27.9%). D-test phenomenon that is
resistance due to MLS-B gene caused by a methylase enzyme
[9]. Out of those 30 (57.7%) were females and 22 (42.3%)
were males. In 45 (86.5%) cases D-test phenomenon was
observed in MRSA while in 7 (13.5%) cases it was also noted
in methicillin sensitive Staph aureus.
A prospective cohort reported from Poland show that the
prevalence and frequency of phenotype of erm gene
encoding enzyme in Staph infections for the years
1991/2010-2011 and 2012 was 59%, 76.9%, 69.7%,
respectively [10]. But after taking remedial action and lawful
use of antibiotic the frequency of D+isolates in 2012, derived
from the culture dramatically decreased to 21.7%.
Erythromycin is a macrolide and clindamycin is a lincosamide
usually treated by clinician as macrolides but chemically and
biochemically clindamycin is different from other macrolides
as it is belongs to lincosamide group. Hence represent two
distinct classes of antibiotic/antibacterial drugs, agents that
inhibit synthesis of protein by inhibiting 50S ribosomal
subunits in ribosomes of SA. In staphylococci, resistance to
both of these agents is caused by a methylase enzyme that
cause mutation in MLS-B antibiotics mediated by erm genes
encoding enzyme.
The sensitivity pattern to D+MRSA in our study
was; vancomycine 94.23%, lanezolid 94.23%, rifampicin
84.23%, fusidic acid 55.77%, doxycyclin 32.59%, levofloxacin
21.15%, gentamycine 13.46%, ciprofloxacin 13.46% [11].
Another study reported that all isolates were found to be
sensitive to vancomycin 100%, the D-test was found to be
positive in 82.4% of the isolates they also suggested that
gentamycin to be used an alternative for treatment of S.
aureus infections with D-test positive, however as per CLSI
aminglycosides alone are not indicated as montherphy and
should be prescribed in combination with another antibiotics
[12]. Their results further showed that vancomycin was the
only antimicrobial agent to be prescribed clinically in infection
caused by D+MRSA.
Conclusion
The sensitivity to vancomycine and lanezolid was 100% in
both D+MSSA and D+MRSA but we did not reported its
sensitivity in cases of D+MSSA to the clinicians to avoid the
misuse of these precious drugs for simple D+MSSA infections
that can easily be treated with beta-lactam antibiotics.
Developed countries have developed stratagies for the use of
vancomycin in clinical practice, an example is USA where they
have designed a computerized structured system in hospital to bound the clinicians starting vancomycin for treating
resistant infections, where the clinicians were supposed to
follow a protocol with clear mention of proper indication of
vancomycin and updating the treatment record in the a
computerized interconnected system to be strictly observed
by the decision makers under management information
system to avoid its misuse.
Irrational, unlawful and inappropriate use of precious
antimicrobial agents like vancomycin and lanizolid is
responsible for emerging resistance in bacteria and also
increase the budget/hospital stay on general public.
It is concluded that a comprehensive strategy using advocacy,
communication social mobilization and CME events can help
in understanding healthcare provider in proper selection of
antibiotics for treatment of various bacterial infections.
There is need for multidisciplinary approached at national,
international and authorities (Medical Teaching Institutions
MTIs) level to develop consensus on lawful and evidence
based use of antibiotic to safeguard the future clinical
challanges.
References
- Prabhu K, Rao S, Rao V (2011) Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. J Lab Physicians. 3(01):025-027.
[Crossref] [Google Scholar] [PubMed]
- Shrestha B, Rana SS (2014) D test: A simple test with big implication for Staphylococcus aureus macrolide-lincosamide-streptograminB resistance pattern. Nepal Med Coll J. 16(1):88-94.
[PubMed] [Google Scholar]
- Yilmaz G, Aydin K, Iskender S, Caylan R, Koksal I (2007) Detection and prevalence of inducible clindamycin resistance in staphylococci. J Med Microbiol. 56(3):342-345.
[Crossref] [Google Scholar] [PubMed]
- Deotale V, Mendiratta DK, Raut U, Narang P (2010) Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Indian J Med Microbiol. 28(2):124-126.
[Crossref][Google Scholar] [PubMed]
- Ajantha GS, Kulkarni RD, Shetty J, Shubhada C, Jain P (2008) Phenotypic detection of inducible clindamycin resistance among Staphylococcus aureus isolates by using the lower limit of recommended inter-disk distance. Indian J Pathol Microbiol. 51(3):376-378.
[Crossref] [Google Scholar] [PubMed]
- Lowy FD (2003) Antimicrobial resistance: The example of Staphylococcus aureus. J Clin Invest. 111(9):1265-1273.
[Crossref] [Google Scholar] [PubMed]
- GMisic M, Cukic J, Vidanovic D, Sekler M, Matic S, et al. (2017) Prevalence of genotypes that determine resistance of staphylococci to macrolides and lincosamides in Serbia. Front Public Health. 5:200.
[Crossref] [Google Scholar] [PubMed]
- Woods CR (2009) Macrolide-inducible resistance to clindamycin and the D-test. Pediatr Infect Dis J. 28(12):1115-1118.
[Crossref] [Google Scholar] [PubMed]
- Szymanek-Majchrzak K, Mlynarczyk A, Bilinska M, Rownicki M, Majchrzak K, et al. (2018) Effect of selective antibiotic pressure on the MLS-B phenotype in methicillin-resistant Staphylococcus aureus strains originating from patients from transplantation wards: 24 years of observations. Transplant Proc. 50(7):2164-2169.
[Crossref] [Google Scholar] [PubMed]
- Ojulong J, Mwambu TP, Joloba M, Bwanga F, Kaddu-Mulindwa DH (2009) Relative prevalence of methicilline resistant Staphylococcus aureus and its susceptibility pattern in Mulago Hospital, Kampala, Uganda. Tanzan J Health Res. 11(3):149-153.
[Crossref] [Google Scholar] [PubMed]
- Shojania KG, Yokoe D, Platt R, Fiskio J, Ma'Luf N, et al. (1998) Reducing vancomycin use utilizing a computer guideline: Results of a randomized controlled trial. J Am Med Inform Assoc. 5(6):554-562.
[Crossref] [Google Scholar] [PubMed]
- Feucht CL, Rice LB (2003) An interventional program to improve antibiotic use. Ann Pharmacother. 37(5):646-651.
[Crossref] [Google Scholar] [PubMed]
Citation: Khan H, Bari F, Basharat T, Khan MB, Saadullah, et al. (2024) Phenotypic Detection of erm Gene Encoding Methylase
for Resistance to Macrolide, Lincosamide-Streptogramin-B (MSL-B) in Methicillin Sensitive and Methicillin Resistant Staph
aureus Infections in a Tertiary Care Hospital of Nowshera. Pediatr Health Res. 9:14.
Copyright: © 2024 Khan H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution
License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source
are credited.

