Research Article - (2016) Volume 2, Issue 1
Neelam Kedia1,2, Leena Kadam1,3, Kushaan Dumasia1 and Balasinor NH1*
1Neuroendocrinology Division, National Institute for Research in Reproductive Health, Parel, Mumbai, India
2Institute for Stem Cell Biology and Regenerative Medicine, National Centre for Biological Sciences, Tata Institute of Fundamental Research, GKVK, Bellary Road, Bangalore, India
3Department of Physiology and of Obstetrics and Gynaecology, School of Medicine, Wayne State University, Detroit, Michigan, USA
*Corresponding Author:
Balasinor NH
Neuroendocrinology Division, National Institute for Research in Reproductive Health
Parel, Mumbai-400012, India
Tel: +912224192025 +912224192140
Fax: +912224139412
E-mail: balasinorn@nirrh.res.in necnirrh@hotmail.com
Received date: January 22, 2016; Accepted date: February 08, 2016; Published date: February 15, 2016
Citation: Kedia N, Kadam L, Dumasia K, et al. Possible Role of Paternal Aberrant Imprinting in Placental Development: A Study in Tamoxifen Treatment Rat Model. J Clin Epigenet. 2016, 2:1. DOI: 10.21767/2472-1158.100014
Copyright: © 2016 Balasinor NH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Genomic imprinting is known to regulate fetal growth and development. Studies from our laboratory have demonstrated that treatment of adult male rats with tamoxifen increased post-implantation loss around mid-gestation. Microarray analysis of resorbing vs. normal embryos revealed aberrant expression of imprinted genes and deregulation of genes associated in placental function. In the present study, expression levels of placental lineage specific genes and imprinted genes implicated in placental development in resorbing embryos sired by tamoxifen treated male rats were evaluated by qPCR. The morphology of these embryos sired was assessed by routine haematoxylin-eosin staining. Flow cytometry was done to confirm the observed results on increase in trophoblast giant cells. Expression of genes responsible for the formation of trophoblast giant cells, spongiotrophoblast and labyrinth was affected in the resorbing embryos. Morphology of the normal and resorbing embryos sired by tamoxifen treated male rats showed increase in giant trophoblast cells along with reduction in spongiotrophoblast and labyrinth trophoblast in the resorbing embryos. The study suggests that aberrant imprinting observed upon tamoxifen treatment of adult male rats affects placental development by altering the bias towards development of one cell lineage, thus abrogating normal placental structures and highlights role for paternal imprinting in placentation.
Keywords
Placental morphology; Genomic imprinting; Trophoblast giant cells; Embryo loss
Abbreviations
ICM: Inner Cell Mass; TE: Trophectoderm; TGC: Trophoblast Giant Cells; EP: Ectoplacental Cone; TS: Trophoblast Stem Cells; POL: Post Implantation Loss; CP: Control Normal Placenta; TP: Treated Normal Placenta; CR: Control Resorbing; TR: Treated Resorbing; Cdx2: Caudal Type Homeobox 2; Bhlh: Basic Helix-Loop-Helix; Hand1: Heart and Neural Crest Derivatives Expressed 1; Stra13: Stimulated by Retinoic Acid 13; Ascl2: Achaete-Scute Family bHLH Transcription Factor 2; Gcm1: Glial Cells Missing-1; Peg3: Paternally Expressed Gene 3; Dcn: Decorin; Peg1/Mest: Paternally Expressed Gene 1; Dlk1: Delta-like 1 Homolog; Cdkn1c: Cyclin-Dependent Kinase Inhibitor 1C
Introduction
Classical nuclear transplantation experiments demonstrated the functional non-equivalence of the two parental pronuclei during embryogenesis and the biasness of paternal genome towards the development of extra-embryonic lineages [1]. This functional non-equivalence is genomic imprinting, whereby a subset of genes is expressed from only one specific parental chromosome. Most imprinted genes are implicated in growth and development, and their parental-specific expression is established through epigenetic modifications in the germ line [2].
In mammals, the placenta mediates the exchange of nutrients between mother and foetus thereby modulating embryo growth. Placenta formation begins with the segregation of cells of the morula into the inner cell mass (ICM) and trophectoderm (TE) [3]. In rodents, after implantation, the mural trophectoderm undergoes differentiation and endoreduplication giving rise to primary trophoblast giant cells (TGC) which invade the uterus. The polar trophectoderm in contact with ICM grows to form the ectoplacental cone (EPC) and the extraembryonic ectoderm, both of which contribute to the formation of the placenta. The extraembryonic ectoderm develops into the trophoblast cells of the chorion layer and gives rise to the labyrinth. The EPC cells divide to form the spongiotrophoblast which is the compact layer of cells sandwiched between the labyrinth and the outer giant cell layer [3]. The cells of the EPC also give rise to secondary TGCs. The TGC invade into the maternal decidua and are thus important for anchoring the placenta, whereas the labyrinth layer is the main site of nutrient exchange. The intermediate spongiotrophoblast layer supports the labyrinth and has been recently implicated to secrete growth factors. Absence and/or defects in either of the 3 layers has been associated with placental dysfunction [4,5].
The genes involved in specification and differentiation of these cells types have been elucidated. Cdx2, a homolog of the Drosophila homeotic caudal (Cad) gene is involved in the segregation of the ICM and TE lineages and its expression is restricted to the TE [6]. The genes, Hand1 and Stra13 are responsible for differentiation of TGC and are exclusively expressed only in these cells of the placenta. Over-expression of either Hand1 or Stra13 in TS cell cultures induces differentiation to TGCs [7]. Gcm1 (Glial cells missing-1) controls cell-fate decisions and its expression is confined to the labyrinth layer (including syncytiotrophoblast cells) in mouse placenta. Gcm1 knockout mice die around embryonic day 10 (E10) due to lack of labyrinth trophoblast [8,9] Ascl2 ( Mash2), a mammalian member of Achaete-Scute family, is essential for spongiotrophoblast formation and negatively affects TGC differentiation. Ascl2 knockout mice die by E10 due to placental defects, which include the absence of spongiotrophoblast, an increased number of TGCs and a failure in labyrinth formation [10-12].
Apart from these, a few imprinted genes have been shown to be important for placental development. The paternally expressed genes, Mest/Peg1, Peg3 both show expression in the trophoblast cells. Peg3 (Paternally expressed gene 3) is expressed by the cells of spongio- and labyrinth trophoblast and secondary giant cells. Peg3 null mice show growth abnormalities and exhibit 25% reduction in placental size [13-15]. Peg1 is expressed in the villous and invasive cytotrophoblast cells of the human placenta [16]. The endothelial cells of the mouse placenta also express Peg1 gene and the null mice show fetal and placental growth retardation [17]. Another paternally expressed gene, Dlk1 (Deltalike 1 homolog) was demonstrated to contribute to mouse placentation [18]. It is involved in endothelial development and formation of the labyrinth [19]. The Dlk1 null mice display greater than 50% perinatal lethality; the surviving mice show severe growth retardation and undergrowth of the placenta [20,21].
The Dcn (decorin) gene is maternally expressed only in the placenta and is implicated to control proliferation, migration and invasiveness of trophoblast cells [22-24]. Cdkn1c (Cyclindependent kinase inhibitor 1C), residing in the Beckwith- Wiedmann cluster, is involved in placental development [25]. Cdkn1c is involved in G1 cell cycle arrest and has been implicated in endoreduplication of TGCs [26]. The Cdkn1c null mice display placentomegaly, with dysplasia of labyrinth and spongiotrophoblast layers [25].
Placental abnormalities account for myriad of developmental problems ranging from intrauterine growth retardation to embryo loss [27-29]. Our laboratory has been working with tamoxifen, a selective estrogen receptor modulator in understanding its effect on male fertility. Oral administration of Tamoxifen to adult male rats at the dose of 0.4 mg/kg/day for 60 days increases pre- and post-implantation loss without affecting the fertilizing ability [30,31]. This embryo loss correlated with the hypomethylation of Igf2-H19 imprint control region (ICR) [32]. Microarray expression studies on the embryos sired by tamoxifen treated rats, revealed an effect on development, growth, metabolism and differentiation associated pathways in the resorbing embryos [33]. Aberrant expression of imprinted genes namely Igf2, Dlk1, Plagl1, Osbpl5, Ascl2, Cdkn1c, Phlda2, Igf2r, Grb10, Gpc3, Tbpba and Sgce was also observed in the resorbing embryos. Down-regulation of cyclins and their dependent kinases and up-regulation of Tgf, Cdkn1a and Cdkn1b suggested an arrest in cell cycle at the G1- phase in the resorbing embryos [34]. Interestingly, preliminary studies on the histology of the resorbing embryos (at embryonic day 20) showed trophoblastic outgrowths [31].
As placental development is crucial for the survival of the embryo, in the present study we further characterize the placental defects in the resorbing embryos obtained from tamoxifen treated rats at E13. We studied the histology in more detail and also expression of transcripts of placental lineage specific marker genes namely Cdx2, Stra13, Gcm1 and Hand1. We also studied expression imprinted genes namely Peg1, Peg3 and Dcn, known to play a role in placental function; in the normal and resorbing embryos obtained from tamoxifen treated rats at E13.
Materials and Methods
Animals and treatment schedule
Randomly bred adult male (75 days old) and female (90 days old) rats of Holtzman strain, maintained at the temperature of 22-23°C, humidity 50-55%, and light: dark cycle of 14:10 h in the Institutes Animal House were used for the study. Food and water were available ad libitum. The study was approved by the Institute’s Animal Ethics Committee.
Tamoxifen citrate tablets (Hetero Drugs Ltd) containing 10 mg of tamoxifen was uniformly suspended in water by sonication and administered orally to adult male rats at a concentration of 0.4 mg/kg/day, five days a week for 60-days via a rat feeding tube [30,31]. Control male rats were administered water only. Thirty male rats per group were used. One week before the end of treatment, the male rats were cohabited with normal cycling female rats in the ratio of 1 male: 2 females. The day of sperm positive vaginal smears were taken as day ‘0’ of gestation. Gravid rats were sacrificed on E13, normal and resorbing embryos were separately processed for histology, flow cytometry and few were snap frozen and stored at -80°C (for RNA extraction). Resorbing embryos were distinguished on the basis of their small size. The percentage of post-implantation loss (POL) was calculated using the following formula. The data was compared by one-way ANOVA (SPSS) and significant differences between groups was computed by the Duncan’s Multiple Range test and p < 0.05 was considered significant.

RNA extraction and real time RT-PCR
RNA extraction: RNA extraction from individual embryos and cDNA preparations was done as described by [35]. RNA was extracted from individual embryos (normal/resorbing) at E13 using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA samples were further purified on the RNeasy mini-kit columns (Qiagen, Hilden, Germany) which also included a step of DNAse digestion. RNA concentration was assessed by measuring absorbance at 260 nm on a spectrophotometer (Shimadzu, Japan). The purity and the integrity of the RNAs were checked by measuring the optical density at 260 and 280 nm followed by electrophoresis on a 1.5% agarose gel stained with 0.01% ethidium bromide (Sigma-Aldrich). 2 μg of RNA was converted to cDNA with random primers using High-Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) as detailed by the manufacturer. Resulting cDNA was diluted 1 in 10, aliquoted and stored at -80°C.
Real-time PCR: Quantitative real-time PCR was performed on an ABI 7900HT Fast Real-time PCR System (Applied Biosystems). Amplification with house-keeping gene 18S rRNA (Eukaryotic 18S rRNA Endogenous Control TaqMan assay from Applied Biosystems) was performed to determine the integrity of the cDNA samples. The relative expression of genes Cdx2, Stra13, Gcm1, Peg1, Peg3, and Dcn (TaqMan Gene Expression Assays from Applied Biosystems) was assessed with respect to housekeeping gene 18S rRNA. The TaqMan Gene Expression Assays are designed and pre-validated so they all have amplification efficiencies averaging 100% (+/- 10%). Biplex ampli?cation reactions were set up in 20 μl volume, containing 2.0 μl of cDNA from normal or resorbing embryos, 1 μl of the respective gene specific primer mix containing FAM-labeled probes, 1 μl of the 18S rRNA primer mix containing VIC-labeled probe and 1x TaqMan Universal PCR Master Mix containing Amp-Erase UNG (Applied Biosystems) according to manufacturer’s instructions with the following thermal cycling conditions: 1 cycle at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were performed in triplicates using MicroAmp fast optical 96-well reaction plates with barcode (Applied Biosystems) and sealed with MicroAmp optical adhesive film (Applied Biosystems). The details of the primer-probe are provided in Table 1. Negative controls were run in which the RNA templates were replaced by nuclease-free water in the reactions. The Ct values remained either undetermined or extremely low compared to gene specific PCR product for the negative control reactions. In the no-RT controls, nuclease-free water was used in place of the reverse transcriptase, no amplification was observed in these reactions. The PCR reactions were set-up using SDS Plate Utility Software Version 2.1 software (Applied Biosystems) and analysis of relative quantitation data was done using RQ Manager 1.2 software (Applied Biosystems).
| Gene name | Gene symbol | Assay ID | Target exon | Amplicon length (bp) | Assay Location | RefSeq ID/Celera |
|---|---|---|---|---|---|---|
| Bhlhb2/Stra13 | basic helix-loop helix domain containing, class B2 | Rn00584155_m1 | 4-5 | 107 | 702 | NM_053328.1 |
| Gcm1 | Glial cells missing homolog 1 (Drosophila) | Rn00820824_g1 | 5-6 | 120 | 733 | NM_017186.2 |
| Cdx2 | Caudal type homeo box 2 | Rn00576694_m1 | 1-2 | 89 | 651 | NM_023963.1 |
| Peg1 | Paternally expressed gene 1 | Rn01500324_m1 | 10-11 | 101 | 1084 | NM_001009617.1 |
| Dcn | Decorin | Rn01503161_m1 | 7-8 | 101 | 915 | NM_024129.1 |
| Peg3 | Paternally expressed 3 | Rn01523805_m1 | 1-2 | 75 | 167 | rCT32052.0 |
Table 1: Details of gene expression assay used for real time RT-PCR primer.
To determine the ΔCt values the Ct values obtained for target gene were normalized to the endogenous control (18S rRNA). The ΔΔCt was calculated using control normal (CN) embryo sample as calibrator (CN1 for E13 cDNA samples). Relative quantitation values (RQ) were obtained using the formula: RQ = 2-ΔΔCt where ΔΔCt = Ct (test sample) - Ct (calibrator) [36]. To determine the level of fold change, Log RQ to base 2 values were calculated. Mean ± SE values were analyzed by one-way ANOVA (GraphPad Prism 5) and significance between groups was determined by Bonferroni multiple range test at P < 0.05. Five sets of normal and resorbing embryos at E13 from control and treated groups were used for the study, such that each set of normal/resorbing embryos were sired by different males. Real-time PCR procedures and analyses follow the MIQE guidelines [37] (Supplementary Table 1).
Real-time quantitative RT-PCR for Hand1 was performed with 1 μl of cDNA per sample using SYBER Green (Bio-Rad Laboratories). The primer sequences used for the PCR for Hand1 were F: 5'-AAGCAAGCGGAAAAGGGAGT-3' and R: 5'-ACTGATTAGCTCCAGCGCC- 3' and for 18S were F: 5’-CCGCAGCTAGGAATAATGGA- 3′and R: 5′ AGTCGGCATCGTTTATGGTC 3′the annealing temperature for both the primers was 60°C. Reactions were incubated at 95°C for 10 min and cycled according to the following parameters: 95°C for 30 s (melt) and 60°C for 1 min (anneal/extend) for a total of 40 cycles. All experiments were performed in duplicates. Negative control and ‘no-RT’ control reactions were performed as described above. Relative quantitation and fold change was calculated as described above.
Histology of post-implantation embryos
Normal and resorbing embryos at E13 were fixed in Bouin’s fixative for 24 h and then dehydrated through ascending concentrations of ethanol, cleared in xylene and embedded in paraffin wax. The entire blocks containing a whole embedded embryo, were serially sectioned into 8 μm sections. Sections were stained with Hematoxylin and Eosin stains, dehydrated and cleared. The stained sections were viewed with AxioScope.A1 microscope (Carl Zeiss). The sections containing the entire placenta and embryopart were selected for morphometric measurements (Figure 1). High-resolution overview images from individual images were generated using Panorama, AxioVision 1.0v Software Module.
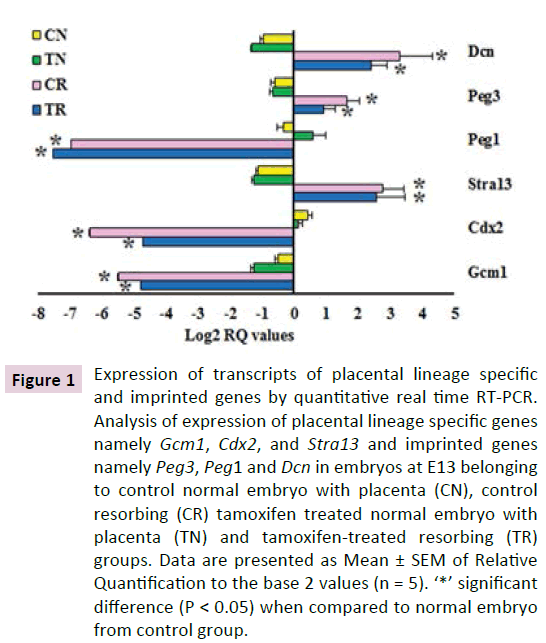
Figure 1 Expression of transcripts of placental lineage specific and imprinted genes by quantitative real time RT-PCR. Analysis of expression of placental lineage specific genes namely Gcm1, Cdx2, and Stra13 and imprinted genes namely Peg3, Peg1 and Dcn in embryos at E13 belonging to control normal embryo with placenta (CN), control resorbing (CR) tamoxifen treated normal embryo with placenta (TN) and tamoxifen-treated resorbing (TR) groups. Data are presented as Mean ± SEM of Relative Quantification to the base 2 values (n = 5). ‘*’ significant difference (P < 0.05) when compared to normal embryo from control group.
Assessment of length of trophoblast layer
The length of the trophoblast layer in three different regions within a microscopic field was measured. Length in four such microscopic fields per section was measured; average length of the trophoblast layer per section was determined and represented as mean ± SD in μm (Figure 1). Three embryos from each group were assessed and the results were analyzed by one-way ANOVA (GraphPad Prism). Significance between groups was determined by Duncan’s Multiple Range test at P < 0.05.
Flow cytometric analysis of placental cells
Gravid rats were sacrificed at E13 and the embryos (both normal and resorbing) were dissected from the uterine covering and collected individually in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich). In case of normal embryos the placental part was dissected and sieved through a 70 μm filter (BD Biosciences) to obtain a single cell suspension. In case of resorbing embryos, the placental part could not be separated hence the entire tissue was used for preparation of single cell suspension. The aliquots of 1 × 106 cells/ml were fixed using chilled 70% alcohol and stored at 4°C.
Alcohol fixed cells were washed twice with 0.1 M Phosphate buffer solution (NaCl 137 mM, KCl 2.7 mM, Na2HPO4 10 mM, KH2PO4 1.8 mM). The cells were sieved through a 70 μm filter (BD Biosciences) again, to remove clumps and obtain a uniform single cell suspension. The cells were suspended in 0.1 M PBS containing 0.1% Nonidet P-40 (NP-40, Roche Diagnostics), 150 μg of RNase solution (Sigma-Aldrich) and 25 μg Propidium Iodide (Sigma-Aldrich), incubated in dark for 30 minutes and analysed by FACS Aria (BD Biosciences). A total of 10,000 events were acquired and analysed ungated using Cell Quest software (FACS diva version 6.1.3). A total of 3 embryos per group, viz control normal placenta (CP), treated normal placenta (TP), control resorbing (CR) and treated resorbing (TR) were analysed in duplicates. Statistical analysis was carried out using the one-way ANOVA (SPSS) and significance between groups was determined by Duncan’s Multiple Range test at P < 0.05.
Results
Post implantation loss observed in embryos sired by tamoxifen treated male rats
The percentage of post implantation loss (POL) in the control group was 3 ± 0.7 whereas in the treatment group it was 10 ± 2 (values represented mean ± SEM).The percentage of POL in treated groups was significantly higher as compared to the control group which also was observed in our previous studies [35].
Expression of transcripts of placental markers and imprinted genes in embryos sired by tamoxifen treated male rats
We studied the expression of Gcm1, Cdx2, Stra13, Hand1, trophoblast differentiation related genes and Peg1, Peg3, and Dcn, imprinted genes (Figure 1). Except for Peg1, the genes did not show any differences in the expression levels in the normal embryos from the control and treatment groups. The transcripts of Gcm1, Cdx2 and Peg1 showed decreased expression in the resorbing embryos from both the control and treated groups when compared to the normal embryos of the respective groups. The transcripts of Stra13, Peg3 and Dcn showed increased expression in the resorbing embryos compared to their respective normal embryos. Interestingly, difference in expression levels was observed between the resorbing embryos from the control and treatment groups. Resorbing embryo from the treatment group had 3.18 folds higher expression of Cdx2 when compared to the resorbing embryos of the control group (Figure 1). Expression of Hand1 transcripts was also increased 7 folds, in resorbing embryos from the treatment group as compared to resorbing embryos from the control group (Figure 2).
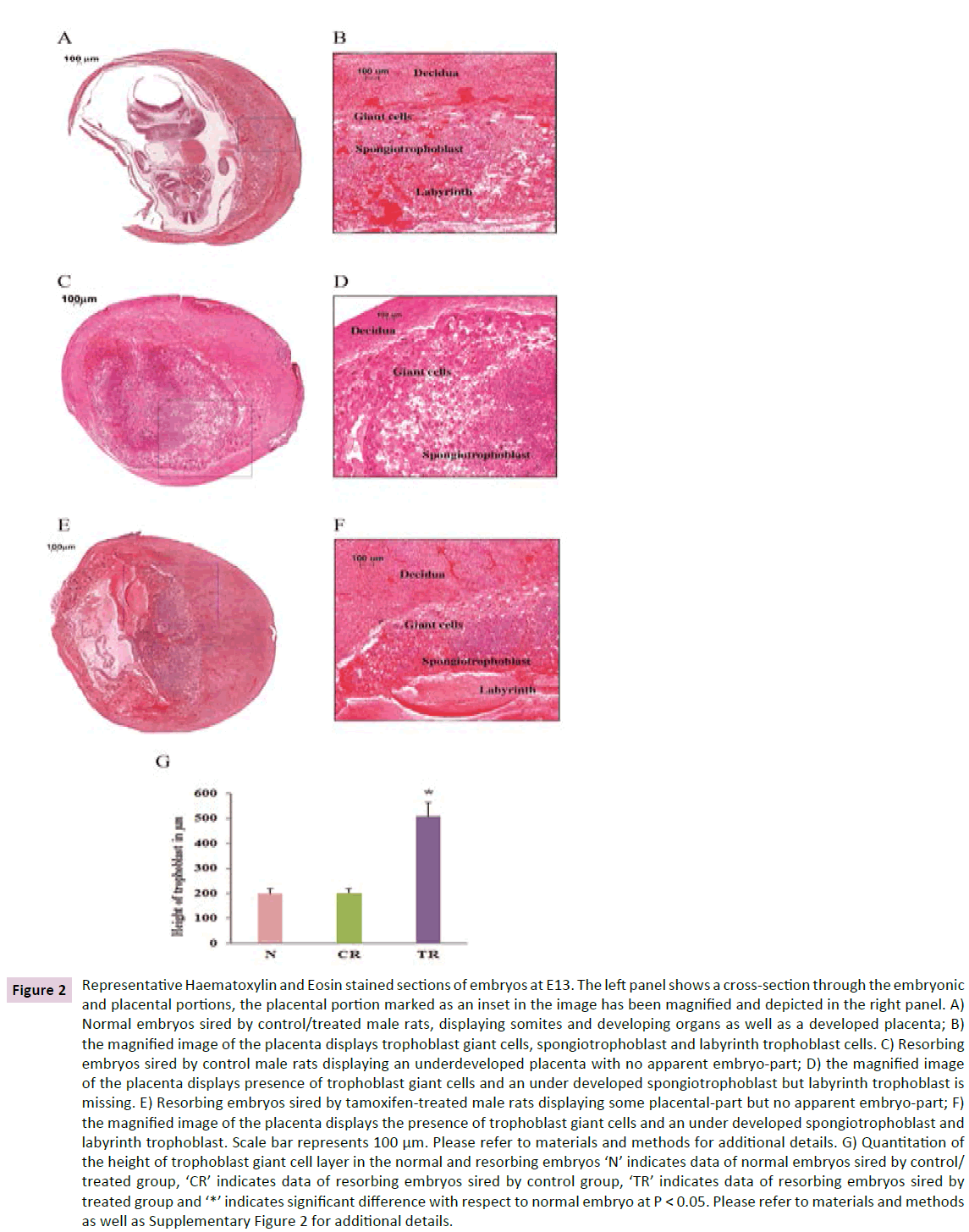
Figure 2 Representative Haematoxylin and Eosin stained sections of embryos at E13. The left panel shows a cross-section through the embryonic and placental portions, the placental portion marked as an inset in the image has been magnified and depicted in the right panel. A) Normal embryos sired by control/treated male rats, displaying somites and developing organs as well as a developed placenta; B) the magnified image of the placenta displays trophoblast giant cells, spongiotrophoblast and labyrinth trophoblast cells. C) Resorbing embryos sired by control male rats displaying an underdeveloped placenta with no apparent embryo-part; D) the magnified image of the placenta displays presence of trophoblast giant cells and an under developed spongiotrophoblast but labyrinth trophoblast is missing. E) Resorbing embryos sired by tamoxifen-treated male rats displaying some placental-part but no apparent embryo-part; F) the magnified image of the placenta displays the presence of trophoblast giant cells and an under developed spongiotrophoblast and labyrinth trophoblast. Scale bar represents 100 μm. Please refer to materials and methods for additional details. G) Quantitation of the height of trophoblast giant cell layer in the normal and resorbing embryos ‘N’ indicates data of normal embryos sired by control/ treated group, ‘CR’ indicates data of resorbing embryos sired by control group, ‘TR’ indicates data of resorbing embryos sired by treated group and ‘*’ indicates significant difference with respect to normal embryo at P < 0.05. Please refer to materials and methods as well as Supplementary Figure 2 for additional details.
The imprinted genes Peg1, Peg3 and Dcn were down regulated 1.4, 1.6 and 1.8 folds respectively in the resorbing embryos of the treatment group when compared to resorbing embryos of control group.
Histology of post-implantation embryos sired by tamoxifen treated male rats
Histology of normal embryos at E13 showed presence of somites and cross sections through the developing organs. The placenta of these embryos showed distinct labyrinth, spongiotrophoblast and giant trophoblast cell layers. However, the resorbing embryos at E13 showed an underdeveloped labyrinth and spongiotrophoblast layers but the TGC layer was found to be increased when compared to the normal embryos (Figure 2).
As the spongiotrophoblast and labyrinth layers were underdeveloped in the resorbing embryos, we could not compare their thickness with the normal embryos. Hence we assessed the length of the TGC layer in normal and resorbing embryos. The length of TGC layer was comparable in normal embryos from control and treated groups. Therefore we combined them for statistical analysis and graphical representation. Interestingly, the resorbing embryos of the treated group consistently showed a thicker TGC layer. The giant cell layer was approximately 2.5 times larger in these embryos when compared to the normal embryos as well as resorbing embryos of the control group (Figure 2). The length of the layer in the resorbing embryos from the control group was comparable to that in the normal embryos.
Flow cytometry for ploidy analysis
Since TGCs are polyploidy cells the observed increase in length of this layer was confirmed by cell cycle analysis using flow cytometry (Figure 3). For comparison, placental cells obtained from normal embryos of both the groups were analysed. A significant decrease in the percentage of cells in S-phase and G2- phase was observed in resorbing embryos from both the group as compared to placental cells from normal embryos. A significant increase in the percentage of polyploid cells (> 4n) was observed in the resorbing embryos of treated group, exclusively. The percentage of the same was comparable in placentae of normal embryos of both groups and the resorbing embryos of the control group. The resorbing embryos of the treated group additionally showed a significantly higher percentage of apoptotic cells when compared to all the other groups.
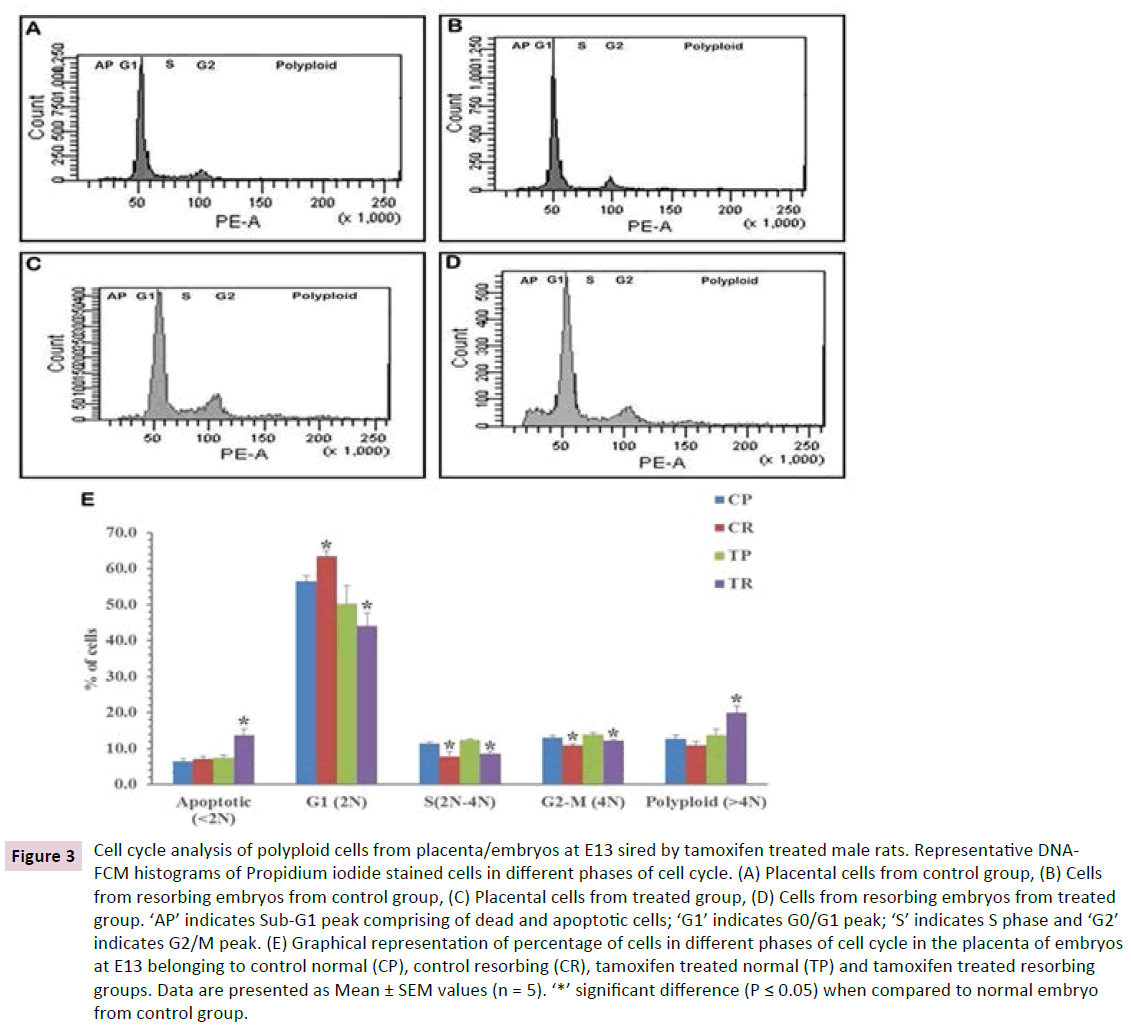
Figure 3 Cell cycle analysis of polyploid cells from placenta/embryos at E13 sired by tamoxifen treated male rats. Representative DNAFCM histograms of Propidium iodide stained cells in different phases of cell cycle. (A) Placental cells from control group, (B) Cells from resorbing embryos from control group, (C) Placental cells from treated group, (D) Cells from resorbing embryos from treated group. ‘AP’ indicates Sub-G1 peak comprising of dead and apoptotic cells; ‘G1’ indicates G0/G1 peak; ‘S’ indicates S phase and ‘G2’ indicates G2/M peak. (E) Graphical representation of percentage of cells in different phases of cell cycle in the placenta of embryos at E13 belonging to control normal (CP), control resorbing (CR), tamoxifen treated normal (TP) and tamoxifen treated resorbing groups. Data are presented as Mean ± SEM values (n = 5). ‘*’ significant difference (P ≤ 0.05) when compared to normal embryo from control group.
Discussion
The transcription factors Cdx2, Hand1, Gcm1, and Stra13 are known to regulate the differentiation and maintenance of the trophoblast cell subtypes [38]. Cdx2, Hand1 and Stra13 expression is known to regulate TGC formation [7,39-42]. Ascl2 is required for maintaining spongiotrophoblast cells [10,12] and Gcm1 for the formation and maintenance of labyrinth [8,9]. We have previously shown down regulation of Ascl2 in resorbing embryos sired by tamoxifen treated males [35]. In the present study, we report aberrant expression of transcripts of Cdx2, Stra13 Gcm1 and Hand1 genes in these resorbing embryos. Down regulation of genes governing placental cell differentiation, points towards the defects in placenta formation.
In addition, we observed aberrant expression of imprinted genes Peg1, Peg3 and Dcn in the resorbing embryo from the tamoxifen treatment group, known to play crucial roles in rodent and human placental development. We had earlier reported hypermethylation of imprinting control regions of Plagl1, Peg3, Peg5, Igf2r, Grb10 and Dlk1-Gtl2 loci in the resorbing embryos sired by tamoxifen treated male rats at E13 which could have elicited aberrant expression of imprinted genes in these embryos [33]. Aberrant imprinting at the Igf2-H19 locus in spermatozoa was observed after tamoxifen treatment rats which correlated with the percentage of post-implantation embryo loss observed in these rats [32]. Recently, hypomethylation at the IGF2-H19 imprint control region was observed in spermatozoa obtained from idiopathic cases of recurrent spontaneous abortions [34]. These studies together show a strong association between deregulation of imprinted gene expression due to paternal imprinting aberrations, and increased incidence of embryo loss in humans and rats.
Appropriate expression of imprinted genes is crucial for placental development therefore we studied histology of the embryos obtained after paternal tamoxifen treatment. The resorbing embryos from both control and tamoxifen treated groups were severely growth retarded in comparison to the normal embryos. Interestingly, the resorbing embryos from tamoxifen-treated group displayed a thick TGC layer. Also, the spongiotrophoblast and labyrinth trophoblast layers of the placenta were either absent or decreased in these embryos. This was confirmed by flow cytometry, where we observed a significant increase in the percentage of polyploid cells in the resorbing embryos of the treatment group in comparison to the other groups.
The transcripts assessed in the present study combined with previously reported microarray studies have shown downregulation of the transcripts of Cdx2, Ascl2, Hand1 and Gcm1 whilst up-regulation of Stra13 transcripts in the resorbing embryos. These genes are involved in the differentiation to different placental cell lineages as mentioned earlier. Formation of TGCs is considered to be a default pathway and disruption of genes essential for formation of spongiotrophoblast and labyrinth has been shown to cause excessive proliferation to the TGC lineage [43]. For example, Ascl2 knockout mice in which the maternal allele is silent, results in embryo death at E10.0 due to placental failure with the mutant placentae showing absence of spongiotrophoblast layer, underdeveloped labyrinth and expansion of TGCs [10,12]. Down regulation of crucial factors required for the development of labyrinth (Gcm1) and spongiotrophoblast (Ascl2) and increase in the factors required for TGC formation (Stra13) may be one of the causes for the trophoblastic giant cell hyperplasia observed in the resorbing embryos of the treated group. It is important to note that, these genes are also affected in the resorbing embryos of the control group but to a lower extent with no observed effects on TGC’s numbers. These embryos also showed up-regulation of Peg3 and Dcn transcripts, to a greater extent compared to those from the treatment group. Since both these genes are involved in autophagy [44] it is possible that the increased TGCs’, which are not functional could have been cleared showing no effect on their numbers.
In the present study, it is intriguing to comprehend, how paternal administration of tamoxifen might be causing structural aberrations in the placenta and leading to resorption of embryos. It is now well known that the paternal epigenome plays a critical role in the development and function of the placenta, which in turn regulates fetal growth [45]. The present study suggests that aberrant imprinting observed upon tamoxifen treatment to adult male rats might be inherited in the placenta. This aberrant methylation may cause developmental bias towards one cell lineage, hence a non-functional placenta causing the resorptions of embryos. The study also point to the involvement of paternal imprinting in placental disorders.
Acknowledgement
We acknowledge the technical assistance of Mr. Suryakant Mandavkar with animal handling, dosage administration and paraffin block sectioning. We are grateful to Mrs. Gayatri Shinde and Mrs. Sushma Khavle for the technical help with flow cytometry, Dr. Sachin Mangale and Mr. Ramesh Kadampal for assistance with imaging sections of embryos with the software Panorama and Mr. Deepak Shelar for technical help. We are grateful to Ms. Anita Kumar and Dr. Mandar Ankolkar for assistance with manuscript writing and submission.
Funding Sources
This study was supported by Board of Research in Nuclear Sciences (BRNS) Department of Atomic Energy (DAE), Government of India with sanction no. 2010/37B/34/BRNS/1431, to N.H.B and Indian Council for Medical Research (ICMR), Government of India, provided Senior Research Fellowship to N.K.
Author’s Contribution
Conceived and designed the experiments: NHB and NK. Analysed the data: NK and LK Wrote the first draft of the manuscript: LK and NK Contributed to the writing of the manuscript: NK, LK and KD. Agree with manuscript results and conclusions: NK, LK, KD and NHB. Jointly developed the structure and arguments for the paper: NK, LK and NHB. Made critical revisions and approved final version: NHB, all authors reviewed and approved of the final manuscript.