Keywords
Pathogenic bacterial; Wound infection; Multidrug resistance; Susceptibility patterns; Gram negative bacteria
Introduction
Wounds infection by bacteria and resistance to common antibiotics are the common post-surgical and medical challenges. Wounds bacterial contamination are the common hospital acquired infections causing more than 80% of the mortality [1]. The most common bacterial genera infecting wounds are Enterococci, Escherichia, Pseudomonas, Klebsiella, Enterobacter, Proteus and Acinetobacter [2,3]. Wounds infection have been a recognized as the most critical problem especially in the presence of foreign materials that increases the risk of serious infection even with relatively small bacterial infection [4]. Nosocomial infection is usually higher in burn patients that correlate with other factors like nature of burn injury, age of patient, extent of injury and burn depth. Other microbial factors such as type, number of organisms, enzymes, toxins production, colonization of the burn wound site, systemic dissemination of the colonizing organisms, have a strong effect on severity of bacterial wound infection [5,6]. As well as, widespread using of vast groups of antibiotics together with the length of time causes a significant development of antibiotic resistance to wound infecting bacteria [7] that subsequently increase the complications and costs of treatment [8].
Thus, the objective of this work was to isolate and characterize various bacterial isolates infecting wounds, and determination of their susceptibility to various common antibiotics.
Materials and Methods
Samples collection
Samples were collected from fifty patient shaving wound infections using a sterile cotton swab from the surface of infected skin. Each sample was inoculated into nutrient agar and McConkey agar media. The plate cultures were incubated for 24 h at 30-37°C [6], bacterial the growth was observed thoroughly.
Identification of isolated pathogenic bacteria
Identification of Gram negative pathogenic bacteria were carried out by API 20E Enterobacteriaceae (BioMérieux Co.). API system has been recognized as a rapid test for bacterial identification. The strips were inoculated by single colony in the suspension media and incubated at 35-37°C for 18-24 h. The results were recorded based on the special chart, three tests were given a code number, the obtained seven digit numbers were expressed to the corresponding organisms according to the API index.
Antimicrobial susceptibility of the pathogenic bacterial isolates
The susceptibility of the recovered bacterial isolates to antibiotics was determined using disc diffusion assay, Kirby-Bauer method [9] as described in of guidelines of the National Committee for Clinical Laboratory Standards [10,11]. Discs of Amikacin 30 μg, Amoxicillin/Clavulanic 20/10 μg, Ceftazidime 30 μg, Ciprofloxacin 5 μg, Colistinsulphate 10 μg, Cefotaxim 30 μg, Cefepime 30 μg, Nitrofurantoin 300 μg, Levofloxacin 5 μg, Rifampicin 5 μg, Sulphamethoxazole/Trimethoprim 25 μg and Tobramycin 10 μg (Oxoid, ThermoFisher, Scientific, USA) were used. After incubation, the diameter of inhibition zone around each disk was measured and the organisms were nominated as sensitive (S), resistant (R) and intermediate resistant (MR).
Results and Discussion
Isolation, identification and prevalence of pathogenic bacterial isolates
Bacterial wound contamination is a serious problem in the hospital and the treatment of wound infections remain a significant concern for surgeons. The risk of developing wound infection depends on the number of bacteria colonies on the wound. The problem has been magnified due to the unrestrained and rapidly spreading resistance to the available array of antimicrobial agents. Fifty cases from wound swabs were collected from different types of wounds, cultured on nutrient and MacConkey media. Forty-one bacterial isolates were isolated from wounds infection out of the total cases. These bacterial isolates were identified based on the API 20E system. From the results, there was 41 cases (82%) recorded as a positive bacterial growth and only 9 samples were recorded as negative bacterial growth. The identified forty-one pathogenic bacterial isolates were belonging to sex genera (Table 1). These genera were Klebsiella pneumoniae (12 isolates; 24%) followed by Pseudomonas fluorescens (10 isolates; 20%), Providencia stuartii (7 isolates; 14%) and Enterobacter cloacae (6 isolates; 12%) and Serratia rubidaea and Citrobacter freundii (3 isolates; 6%). Similarly, high percentage of microbial growth was reported by Mama et al. 91.6% of culture was positive microbial growth and 12.7% had no bacterial growth. Coincident results for K. pneumonia (24%), P. fluorescens (20%), P. stuartii (14%) and E. cloacae (12%), S. rubidaea and C. freundii (6%) were reported [12]. Among the recovered isolates, Pseudomonas species were the most common isolates (48.9%) followed by Citrobacter spp (13.3%), Enterobacter spp (11.1%), Proteus vulgaris (6.6%), Klebsiella spp (2.2%) and Serratia rubidia (2.2%) infecting wounds [12].
| Bacterial Isolates |
Total No. |
Percentage % |
| Klebsiella pneumonia |
12 |
24% |
| Pseudomonas fluorescence |
10 |
20% |
| Providencia stuartii |
7 |
14% |
| Enterobacter cloacae |
6 |
12% |
| Serratia rubidaea |
3 |
6% |
| Citrobacter freundii |
3 |
6% |
Table 1 The different species of bacteria isolated from wound infection.
Antibiotic susceptibility of the pathogenic bacterial isolates
The antibiotic sensitivity of isolated bacterial strains was carried out by Kirby-Bauer disk diffusion assay against 12 antibiotics (Table 2a). Out of the forty-one bacterial isolates, Pseudomonas fluorescens was a completely resistance to all the tested antibiotics. Five pathogenic bacterial isolates belong to the genera Klebsiella, Pseudomonas, Providencia, Enterobacter and Serratia showed a strong resistance to the tested antibiotics by about 91.67%. Seven bacterial isolates belong to Klebsiella pneumoniae (No. 9), Pseudomonas fluorescens (No. 16, 19, 21, 22) and Citrobacter freundii (No. 39, 40) showed 83.33% resistance to all tested antibiotics. Whereas four pathogenic bacterial isolates of Gram negative bacilli namely; K. pneumoniae (No. 3, 12), P. fluorescens (No. 13) and E. cloacae (No. 30) showing 75% resistance to the utilized antibiotic. Four bacterial isolates belonging to K. pneumoniae (No. 6), P. stuartii (No. 28, 29) and E. cloacae (No. 34) showed 66.67% resistance to the different tested antibiotics. While three bacterial isolates belong to Gram negative bacilli namely K. pneumoniae (No. 7), P. fluorescens (No. 20) and P. stuartii (No. 24) showed 58.3% resistance to the tested antibiotics. Also, three pathogenic bacterial isolates K. pneumonia (No. 10), E. cloacae (No. 33) and C. freundii (No. 90) showed 50% resistance to the tested antibiotic. Three pathogenic bacterial isolates K. pneumoniae (No. 4, 5) and P. stuartii (No. 27) showed 41.67% resistance to the different tested antibiotics and six bacterial isolates namely K. pneumoniae (No. 8), P. fluorescens (No. 18), P. stuartii (No. 25, 26) and S. rubidaea (No. 38) showed 33.33% resistance. In addition, K. pneumoniae (No. 11), E. cloacae (No. 35) and S. rubidaea (No. 37) showed 25% resistance to the tested antibiotics. While E. cloacae (No. 32) and S. rubidaea (36) showed 16.67% antibiotic resistance.
| Isolate No. |
Species of bacteria |
Mean inhibition zone (mm)\Reaction to antibiotics |
species Resistance
%(IR+R) |
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
| Susceptibility breakpoint |
| AK ≥ 17 |
AMC
≥ 18 |
CAZ
≥ 18 |
CIP
≥ 21 |
CT
≥ 11 |
CTX
≥ 23 |
F
≥ 17 |
FEP
≥ 18 |
LEV
≥ 17 |
RD
≥ 20 |
SXT
≥ 16 |
TOB
≥ 15 |
| 1 |
Klebsiella pneumoniae |
6\R |
7\R |
6\R |
6\R |
13\S |
6\R |
6\R |
6\R |
6\R |
7\R |
6\R |
6\R |
91.6 |
| 2 |
Klebsiella pneumoniae |
20\S |
19\S |
17\IR |
31\S |
13\S |
8\R |
21\S |
10\R |
27\S |
12\R |
6\R |
15\S |
33.3 |
| 3 |
Klebsiella pneumoniae |
6\R |
6\R |
6\R |
19\IR |
13\S |
6\R |
14\R |
6\R |
16\IR |
7\R |
6\R |
6\R |
75 |
| 4 |
Klebsiella pneumoniae |
21\S |
17\IR |
10\R |
25\S |
13\S |
6\R |
16\IR |
10\R |
21\S |
9\R |
6\R |
17\S |
41.6 |
| 5 |
Klebsiella pneumoniae |
23\S |
18\S |
11\R |
25\S |
12\S |
6\R |
16\IR |
10\R |
21\S |
9\R |
6\R |
16\S |
41.6 |
| 6 |
Klebsiella pneumoniae |
6\R |
18\S |
12\R |
22\S |
10\R |
6\R |
16\IR |
11\R |
20\S |
7\R |
6\R |
6\R |
66.6 |
| 7 |
Klebsiella pneumoniae |
6\R |
20\S |
14\R |
22\S |
10\R |
16\IR |
15\IR |
7\R |
20\S |
7\R |
6\R |
6\R |
58.3 |
| 8 |
Klebsiella pneumoniae |
19\S |
14\IR |
20\S |
21\S |
17\S |
6\R |
18\S |
12\R |
20\S |
9\R |
6\R |
20\S |
33.3 |
| 9 |
Klebsiella pneumoniae |
6\R |
9\R |
6\R |
6\R |
12\S |
13\R |
9\R |
17\IR |
6\R |
7\R |
6\R |
6\R |
83.3 |
| 10 |
Klebsiella pneumoniae |
19\S |
8\R |
6\R |
18\IR |
12\S |
7\R |
17\S |
10\R |
24\S |
9\R |
6\S |
12\R |
50 |
| 11 |
Klebsiella pneumoniae |
20\S |
9\R |
20\S |
25\S |
11\S |
25\S |
17\S |
21\S |
25\S |
8\R |
8\R |
14\IR |
25 |
| 12 |
Klebsiella pneumoniae |
15\IR |
13\R |
14\R |
6\R |
12\S |
6\R |
18\S |
8\R |
7\R |
11\R |
6\R |
6\R |
75 |
Table 2a Antimicrobial susceptibility patterns of the pathogenic Gram negative bacilli against different antibiotics by disc diffusion method. R=Resistant, MR=Moderate, Resistant S=Sensitivity, AK (30 μg)=Amikacin, AMC(30 μg)=Amoxicillin/Clavulanic acid, CAZ(30 μg)=Ceftazidim, CIP(5 μg)=Ciprofloxacin, F(300 μg)=Nitrofurantoin, CT(10 μg)=Colistinsulphate, CTX(30 μg)=Cefotaxim, FEP(30 μg)=Cefepime, SXT(25 μg)=Sulphamethoxazole/Trimethoprim, LEV(5 μg)=Levofloxacin, RD(5 μg)=Rifampicin, TOB(10 μg)=Tobramycin.
The frequency antibiotic resistance of K. pneumonia isolates was summarized in Figure 1. K. pneumoniae isolate No. 1 was only sensitive to Colistin Sulphate, with moderate resistant to the other experimented antibiotics, while, isolate No. 2 of K. pneumoniae was sensitive to seven antibiotics (Amikacin, Amoxicillin/Clavulanic acid, Ciprofloxacin, Colistin Sulphate, Nitrofurantoin, Levofloxacin and Tobramycin); with moderate resistant to Ceftazidim, Isolate No. 3 of K. pneumoniae was sensitive to Colistin Sulphate and moderately resistant to Ciprofloxacin and Levofloxacin. K. pneumoniae (No. 4) isolate was sensitive to Amikacin, Ciprofloxacin, Colistin Sulphate, Levofloxacin and Tobramycin with moderate resistance to Amoxicillin/Clavulanic acid and Nitrofurantoin. K. pneumoniae (No. 5) isolate was sensitive to six types of antibiotics (Amikacin, Amoxicillin/Clavulanic acid, Ciprofloxacin, Colistin Sulphate, Levofloxacin and Tobramycin) and moderate resistant to Nitrofurantoin. K. pneumoniae (No. 6, 7) isolates was sensitive (Amoxicillin/Clavulanic acid, Ciprofloxacin and Levofloxacin) while K. pneumoniae (No. 6) moderate resistant against Nitrofurantoin and K. pneumoniae (No. 7) moderately resistant against Cefotaxim and Nitrofurantoin. K. pneumoniae (No. 8) isolate was sensitive to seven types of antibiotics (Amikacin, Ceftazidim, Ciprofloxacin, Colistin Sulphate, Nitrofurantoin, Levofloxacin and Tobramycin) and moderately resistant against Amoxicillin/Clavulanic acid. While K. pneumoniae (No. 9) isolate was sensitive only to Colistin Sulphate and moderately resistant against Cefepime. Also K. pneumoniae (No. 10) isolate was sensitive to (Amikacin, Colistin Sulphate, Nitrofurantoin, Levofloxacin and Sulphamethoxazole/Trimethoprim) and moderately resistant against Ciprofloxacin. K. pneumoniae (No. 11) isolate was sensitive to eight types of antibiotics (Amikacin, Ceftazidim, Ciprofloxacin, Colistin Sulphate, Cefotaxim, Nitrofurantoin, Cefepime and Levofloxacin) while moderately resistant against Tobramycin. K. pneumoniae (No. 12) isolate was sensitive to Colistin Sulphate and Nitrofurantoin with moderate resistant to Amikacin.
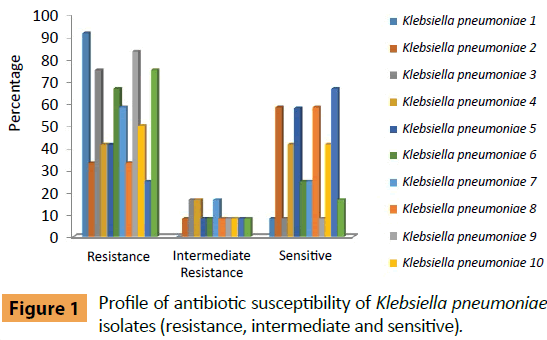
Figure 1: Profile of antibiotic susceptibility of Klebsiella pneumoniae isolates (resistance, intermediate and sensitive).
Pseudomonas fluorescens (No. 13) was sensitive to (Colistin Sulphate, Cefepime and Tobramycin) and moderately resistant against Amikacin, Ceftazidim, Ciprofloxacin and Rifampicin (Figure 2). While P. fluorescens (No. 14) was resistant for all antibiotics. P. fluorescens (No. 15) was sensitive to Colistin Sulphate only and moderately resistant against two types of antibiotics Amoxicillin/Clavulanic acid and Levofloxacin. P. fluorescens (No. 16) was sensitive to Amikacin, Colistin sulphate and moderately resistant to Ciprofloxacin. Whereas P. fluorescens (No. 17) was sensitive to Colistin sulphate and moderately resistant to Nitrofurantoin. P. fluorescens No. 18 was sensitive to Amikacin, Amoxicillin/Clavulanic acid, Ciprofloxacin, Nitrofurantoin, Cefepime, Levofloxacin, Rifampicin and Sulphamethoxazole/Trimethoprim, while it moderately resistant to Ceftazidim and Cefotaxim. Also P. fluorescens No. 19 was sensitive to Colistin Sulphate and Tobramycin. P. fluorescens No. 20 was sensitive to Amikacin, Amoxicillin\Clavulanic acid, Ceftazidim, Colistin Sulphate and Nitrofurantoin, while P. fluorescens No. 21 was sensitive to Colistin Sulphate and Nitrofurantoin and finally P. fluorescens No. 22 was sensitive to Ciprofloxacin and Levofloxacin.
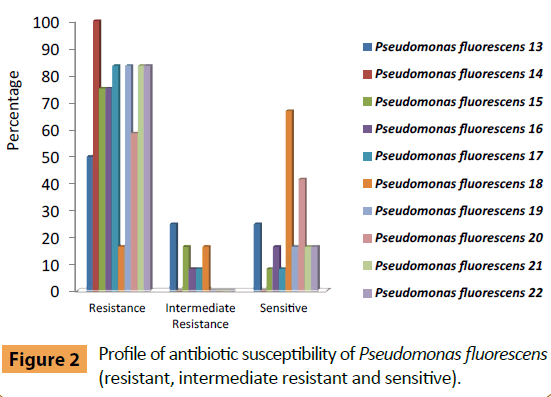
Figure 2: Profile of antibiotic susceptibility of Pseudomonas fluorescens (resistant, intermediate resistant and sensitive).
Providencia stuartii (No. 23) was sensitive to Colistin Sulphate and moderately resistant to Amikacin, Amoxicillin/Clavulanic acid, Levofloxacin and Rifampicin (Figure 3). P. stuartii (No. 24) was sensitive to Amikacin, Ciprofloxacin, Levofloxacin, Sulphamethoxazole/Trimethoprim and Tobramycin without appearance resistant to other antibiotics. While P. stuartii (No. 25) was sensitive to eight types of antibiotics (Amikacin, Ceftazidim, Ciprofloxacin, Colistin Sulphate, Cefotaxim, Cefepime, Levofloxacin and Sulphamethoxazole/Trimethoprim) and moderately resistant to Tobramycin. P. stuartii (No. 26) was sensitive to Amikacin, Ceftazidim, Ciprofloxacin, Cefotaxim, Cefepime, Levofloxacin, Sulphamethoxazole/Trimethoprim and Tobramycin. P. stuartii (No. 27) was sensitive to Amikacin, Ceftazidim, Ciprofloxacin, Cefepime, Levofloxacin, Sulphamethoxazole/Trimethoprim and Tobramycin and moderately resistant to Amoxicillin/Clavulanic acid and Cefotaxim. P. stuartii (No. 28) was sensitive to Amikacin, Ciprofloxacin, Colistin Sulphate and Levofloxacin) and moderately resistant to Nitrofurantoin and Tobramycin. P. stuartii (No. 29) was sensitive to Ciprofloxacin, Colistin Sulphate, Nitrofurantoin and Levofloxacin and moderately resistant to Amikacin.
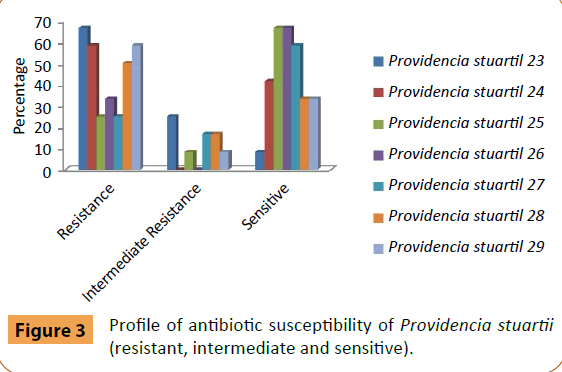
Figure 3: Profile of antibiotic susceptibility of Providencia stuartii (resistant, intermediate and sensitive).
Enterobacter cloacae (No. 30) was sensitive to (Amikacin and Amoxicillin/Clavulanic acid) and moderately resistant against Levofloxacin (Figure 4). E. cloacae (No. 31) was sensitive to only for Colistin Sulphate and without moderately resistant against antibiotics. Also E. cloacae (No. 32) were sensitive to (Amikacin, Ceftazidim, Colistin Sulphate, Cefotaxim, Cefepime, Levofloxacin, Sulphamethoxazole/Trimethoprim and Tobramycin) and moderately resistant against Amoxicillin/Clavulanic acid and Ciprofloxacin. E. cloacae (No. 33) were sensitive to (Ceftazidim, Colistin Sulphate, Levofloxacin and Sulphamethoxazole/Trimethoprim). While moderately resistant against Amoxicillin/Clavulanic acid and Ciprofloxacin. E. cloacae (No. 34) were sensitive to (Colistin Sulphate and Tobramycin) and moderately resistant against Ceftazidim and Rifampicin. Finally, E. cloacae (No. 35) were sensitive to Amikacin, Ceftazidim, Colistin Sulphate, Cefepime and Tobramycin and moderately resistant to Ciprofloxacin, Cefotaxim, Levofloxacin and Rifampicin.
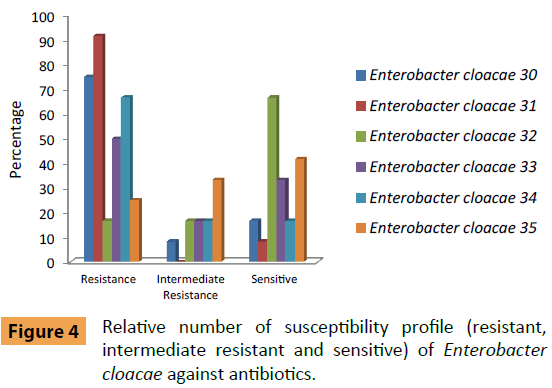
Figure 4: Relative number of susceptibility profile (resistant, intermediate resistant and sensitive) of Enterobacter cloacae against antibiotics.
Serratia rubidaea (No. 36) was sensitive to Amikacin, Amoxicillin/Clavulanic acid, Ceftazidim, Ciprofloxacin, Colistin Sulphate, Cefotaxim, Nitrofurantoin, Cefepime, Levofloxacin and Tobramycin, while moderately resistant to Sulphamethoxazole/ Trimethoprim (Figure 5). S. rubidaea (No. 37) was sensitive to Amikacin, Amoxicillin/Clavulanic acid, Ceftazidim, Colistin Sulphate, Cefotaxim, Nitrofurantoin, Cefepime, Levofloxacin and Tobramycin and moderately resistant to Ciprofloxacin. S. rubidaea (No. 38) was sensitive to Amikacin, Amoxicillin/Clavulanic acid, Ceftazidim, Colistin Sulphate, Cefotaxim, Cefepime, Levofloxacin and Tobramycin) and moderately resistant to Ciprofloxacin and Nitrofurantoin.
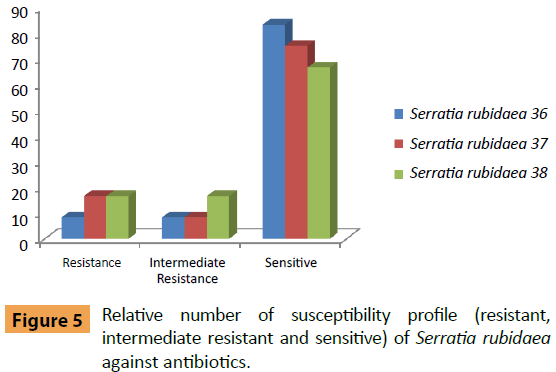
Figure 5: Relative number of susceptibility profile (resistant, intermediate resistant and sensitive) of Serratia rubidaea against antibiotics.
Citrobacter freundii(No. 39) was sensitive to Colistin Sulphate and Levofloxacin and moderately resistant to Ciprofloxacin and Nitrofurantoin (Figure 6). C. freundii(No. 40) was sensitive to Amikacin and Colistin Sulphate while moderately resistant to Amoxicillin/Clavulanic acid and Nitrofurantoin. C. freundii (No. 41) was sensitive to Amikacin, Ceftazidim, Colistin Sulphate, Nitrofurantoin, Sulphamethoxazole/Trimethoprim and Tobramycin and moderately resistant to Amoxicillin/Clavulanic acid.
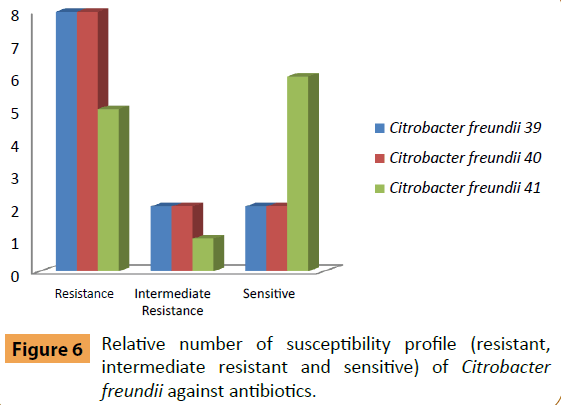
Figure 6: Relative number of susceptibility profile (resistant, intermediate resistant and sensitive) of Citrobacter freundii against antibiotics.
Antimicrobial susceptibility pattern of bacterial isolates was tested against selected 12 antibiotics. From Table 2b the results obtained showed that the bacterial isolates varied in their susceptibility to all the antibiotics and showed that maximum sensitivity for Colistin Sulphate (78.1%), both Amikacin and Levofloxacin (53.7%), Ciprofloxacin (46.3%), Tobramycin (39.1%), Ceftazidim (31.7%), Nitrofurantoin (29.3%), both Amoxicillin/Clavulanic acid and Cefepime (24.4%), Sulphamethoxazole/Trimethoprim (21.9%), Cefotaxim (17.1%) and Rifampicin (2.4%). Whereas bacterial isolates was resistant to Rifampicin (90.2%), Sulphamethoxazole/Trimethoprim (75.6%), Cefotaxim (73.2%), Cefepime (70.7%), Ceftazidim (65.9%), bacterial isolates was resistant to both (Amoxicillin/Clavulanic acid and Tobramycin) (53.7%), Nitrofurantoin (48.8%), Ciprofloxacin (36.6%), both (Cefotaxim and Sulphamethoxazole/Trimethoprim) (34.1%) and bacterial isolates was resistant to Colistin Sulphate (22%). The other results of tested Gram negative bacteria are listed in Table 2c.
| Isolate No. |
Species of bacteria |
Mean inhibition zone (mm)\Reaction to antibiotics |
species Resistance
%(IR+R) |
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
| Susceptibility breakpoint |
| AK ≥ 17 |
AMC
≥ 18 |
CAZ
≥ 18 |
CIP
≥ 21 |
CT
≥ 11 |
CTX ≥ 23 |
F ≥ 17 |
FEP
≥ 18 |
LEV
≥ 17 |
RD
≥ 20 |
SXT
≥ 16 |
TOB
≥ 15 |
| 13 |
Pseudomonas fluorescens |
15\ IR |
6\R |
11\R |
18\ IR |
15\S |
6\R |
6\R |
20\S |
12\R |
18\ IR |
6\R |
17\S |
75 |
| 14 |
Pseudomonas fluorescens |
6\R |
6\R |
10\R |
15\R |
9\R |
6\R |
6\R |
14\R |
6\R |
6\R |
6\R |
6\R |
100 |
| 15 |
Pseudomonas fluorescens |
11\R |
17\ IR |
6\R |
6\R |
13\S |
7\R |
8\R |
7\R |
16\ IR |
8\R |
6\R |
6\R |
91.67 |
| 16 |
Pseudomonas fluorescens |
19\S |
6\R |
6\R |
16\ IR |
14\S |
6\R |
6\R |
6\R |
8\R |
11\R |
6\R |
10\R |
83.33 |
| 17 |
Pseudomonas fluorescens |
6\R |
6\R |
6\R |
6\R |
13\S |
6\R |
15\ IR |
6\R |
6\R |
7\R |
6\R |
6\R |
91.67 |
| 18 |
Pseudomonas fluorescens |
18\S |
21\S |
17\ IR |
26\S |
6\R |
21\ IR |
21\S |
18\S |
18\S |
29\S |
24\S |
12\R |
33.33 |
| 19 |
Pseudomonas fluorescens |
14\R |
6\R |
9\R |
10\R |
12\S |
6\R |
6\R |
9\R |
7\R |
9\R |
6\R |
15\S |
83.33 |
| 20 |
Pseudomonas fluorescens |
20\S |
18\S |
21\S |
6\R |
11\S |
6\R |
18\S |
9\R |
8\R |
10\R |
6\R |
11\R |
58.33 |
| 21 |
Pseudomonas fluorescens |
6\R |
6\R |
6\R |
6\R |
12\S |
7\R |
17\S |
6\R |
6\R |
7\R |
6\R |
6\R |
83.33 |
| 22 |
Pseudomonas fluorescens |
6\R |
6\R |
12\R |
28\S |
6\R |
6\R |
7\R |
9\R |
25\S |
6\R |
6\R |
6\R |
83.33 |
| 23 |
Providencia stuartii |
15\ IR |
16\ IR |
6\R |
6\R |
13\S |
6\R |
9\R |
6\R |
15\ IR |
11\R |
6\R |
6\R |
91.67 |
| 24 |
Providencia stuartii |
22\S |
6\R |
6\R |
30\S |
8\R |
6\R |
10\R |
6\R |
34\S |
14\R |
22\S |
18\S |
58.33 |
| 25 |
Providencia stuartii |
20\S |
10\R |
18\S |
24\S |
18\S |
23\S |
9\R |
20\S |
21\S |
9\R |
17\S |
13\ IR |
33.33 |
| 26 |
Providencia stuartii |
18\S |
12\R |
26\S |
22\S |
6\R |
28\S |
8\R |
25\S |
24\S |
11\R |
21\S |
19\S |
33.33 |
| 27 |
Providencia stuartii |
22\S |
14\ IR |
21\S |
30\S |
8\R |
21\ IR |
11\R |
25\S |
26\S |
8\R |
20\S |
16\S |
41.67 |
| 28 |
Providencia stuartii |
19\S |
7\R |
6\R |
25\S |
13\S |
7\R |
16\ IR |
13\R |
22\S |
6\R |
6\R |
13\ IR |
66.67 |
| 29 |
Providencia stuartii |
15\ IR |
6\R |
6\R |
27\S |
15\S |
7\R |
19\S |
10\R |
22\S |
6\R |
6\R |
7\R |
66.67 |
Table 2b Antimicrobial susceptibility patterns of the pathogenic gram negative bacilli against different antibiotics by disc diffusion method.
| Isolate No. |
Species of bacteria |
Mean inhibition zone (mm)\Reaction to antibiotics |
Species Résistance
%(IR+R) |
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
| Susceptibility breakpoint |
| AK ≥ 17 |
AMC
≥ 18 |
CAZ
≥ 18 |
CIP
≥ 21 |
CT
≥ 11 |
CTX
≥ 23 |
F
≥ 17 |
FEP
≥ 18 |
LEV
≥ 17 |
RD
≥ 20 |
SXT
≥ 16 |
TOB
≥ 15 |
| 30 |
Enterobacter cloacae |
17\S |
18\S |
6\R |
15\R |
6\R |
14\R |
12\R |
11\R |
15\IR |
10\R |
6\R |
8\R |
75 |
| 31 |
Enterobacter cloacae |
6\R |
6\R |
6\R |
6\R |
12\S |
6\R |
6\R |
6\R |
6\R |
6\R |
6\R |
6\R |
91.6 |
| 32 |
Enterobacter cloacae |
24\S |
16\IR |
24\S |
17\IR |
17\S |
26\S |
11\R |
28\S |
21\S |
7\R |
16\S |
17\S |
16.6 |
| 33 |
Enterobacter cloacae |
6\R |
17\IR |
32\S |
18\IR |
16\S |
6\R |
6\R |
6\R |
22\S |
12\R |
18\S |
8\R |
50 |
| 34 |
Enterobacter cloacae |
13\R |
6\R |
16\IR |
15\R |
14\S |
6\R |
6\R |
12\R |
12\R |
17\IR |
6\R |
15\S |
66.6 |
| 35 |
Enterobacter cloacae |
18\S |
6\R |
20\S |
16\IR |
15\S |
16\IR |
6\R |
22\S |
16\IR |
17\IR |
6\R |
18\S |
25 |
| 36 |
Serratia rubidaea |
23\S |
18\S |
32\S |
22\S |
14\S |
34\S |
29\S |
30\S |
24\S |
14\R |
14\IR |
15\S |
16.67 |
| 37 |
Serratia rubidaea |
21\S |
24\S |
25\S |
18\ IR |
13\S |
23\S |
17\S |
30\S |
18\S |
8\R |
6\R |
20\S |
25 |
| 38 |
Serratia rubidaea |
23\S |
22\S |
29\S |
20\ IR |
15\S |
32\S |
16\ IR |
30\S |
21\S |
11\R |
6\R |
15\S |
33.33 |
| 39 |
Citrobacter freundii |
6\R |
6\R |
6\R |
18\ IR |
11\S |
7\R |
15\ IR |
7\R |
17\S |
8\R |
6\R |
6\R |
83.33 |
| 40 |
Citrobacter freundii |
17\S |
16\ IR |
13\R |
6\R |
11\S |
6\R |
16\ IR |
7\R |
6\R |
8\R |
6\R |
11\R |
83.33 |
| 41 |
Citrobacter freundii |
20\S |
16\ IR |
20\S |
6\R |
15\S |
11\R |
21\S |
13\R |
6\R |
7\R |
18\S |
19\S |
50 |
Table 2c Antimicrobial susceptibility patterns of the pathogenic gram negative bacilli against different antibiotics by disc diffusion method.
In our study, Klebsiella pneumoniae (24%) followed by Pseudomonas fluorescens (20%), Providencia stuartii (14%) and Enterobacter cloacae (12%). Both Serratia rubidaea and Citrobacter freundii (6%) with agreement the results were reported [12]. Pseudomonasspecies was found to be the most common isolate (48.9%) followed by Citrobacter braakii (13.3%), Enterobacter spp. (11.1%), Proteus vulgaris (6.66%), Klebsiella spp. (2.22%) and Serratia rubidia (2.22%). As well as, Klebsiella pneumoniae was (83.3%) sensitive to Colistin Sulphate, (66.7%) in Levofloxacin, (58.3%) in Ciprofloxacin, (50%) in Amikacin, (41.7%) in Nitrofurantoin, (33.3%) in both Amoxicillin/Clavulanic acid and Tobramycin. Also (16.6%) in Ceftazidim, finally (8.3%) in both Cefsotaxim and Sulphamethoxazole/Trimethoprim similar to the report in Goswamiet al. While the Klebsiella pneumoniae was (100%) resistance to Rifampicin, (91.7%) in Sulphamethoxazole/ Trimethoprim, (83.3%) in both (Cefotaxim and Cefepime), (75%) in Ceftazidim, (58.3%) in Tobramycin, (50%) in Amoxicillin/Clavulanic acid, (41.7%) in Amikacin, (25%) in both of (Ciprofloxacin, Nitrofurantoin and Levofloxacin) and (16.6%) in Colistin Sulphate that agree with Mamaet al.Similar results were reported for sensitivity and resistance of Gram negative bacteria against selected antibiotics (Figure 7).
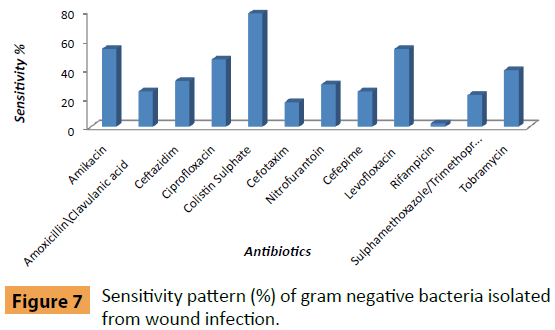
Figure 7: Sensitivity pattern (%) of gram negative bacteria isolated from wound infection.
Conclusion
In conclusion, wound infection by pathogenic bacteria and increasing antibiotics resistance are of the most serious health threats facing the patients, especially diabetic foot patients. Thus, the objective of this work was to isolate and identify pathogenic bacteria infecting wounds. Antimicrobial susceptibility of the isolated pathogenic bacteria to different antibiotics covering all mode of action of antibiotics was conducted.
References
- Manikandan C, Amsath A (2013) Antibiotic susceptibility of bacterial strains isolated from wound infection patients in Pattukkottai, Tamilnadu, India. International Journal of Current Microbiology and Applied Sciences 2: 195-203.
- Gautam R, Acharya A, Nepal HP, Shrestha S (2013) Antibiotic susceptibility pattern of bacterial isolates from wound infection in Chitwan Medical College Teaching Hospital, Chitwan, Nepal. International Journal of Biomedical and Advance Research pp: 248-252.
- Tayfour MA, Al-Ghamdi SM, Al- Ghamdi AS (2005) Surgical wound infections in King Fahad Hospital at Al-Baha. Saudi Med J 26: 1305-1307.
- Rubin RH (2006) Surgical wound infection: epidemiology, pathogenesis, diagnosis and management. BMC Infect Dis 6: 171-172.
- Pruitt BA, McManus AT, Kim SH, Goodwin CW (1998) Burn wound infections: current status. World J Surg 22: 135-145.
- Pruitt BA, Colonel MC, McManus AD (1984) Opportunistic infections in severely burnt patients. Am J Med 76: 146-154.
- Sani RA, Garba SA, Oyewole OA, Ibrahim A (2012) Antibiotic resistance profile of gram positive bacteria isolated from wound infections in Minna, Bida, Kontagora and Suleja Area of Niger State. Journal of Health Sciences 2: 19-22.
- Anguzu J, Olila D (2007) Drug sensitivity patterns of bacterial isolates from septic post-operative wounds in a regional referral hospital in Uganda. Afr Health Sci 7: 148-154.
- Bauer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45: 493-496.
- National Committee for Clinical Laboratory Standards (2005) Performance standards for antimicrobial susceptibility testing. 15th Informational Supplement (M100-S15), Wayne, PA, USA.
- National Committee for Clinical Laboratory Standards (NCCLS) (2000) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS document M7-A5, Wayne, PA, USA.
- Alwan MJL, Hamzah AM (2011) Bacterial isolation from burn wound infections and studying their antimicrobial susceptibility. Kufa Journal for veterinary medical sciences 2: 121-131.







