- (2006) Volume 7, Issue 6
Anil Bagul1, Sudeep Pushpakom2, James Boylan2, William Newman2, Ajith K Siriwardena1
1HPB Unit, Department of Surgery, Manchester Royal Infirmary; 2Academic Department of Medical Genetics, University of Manchester. Manchester, United Kingdom
Received: 02 August 2006 Accepted: 11 September 2006
Context Release of genomic DNA into plasma as a result of necrotic and apoptotic pathways is a feature of a range of human tumours. Severe acute pancreatitis is characterized by inflammation but may also be associated with accelerated apoptotic and necrotic pathways. Objectives This study uses quantitative realtime PCR to measure free circulating DNA in patients with severe acute pancreatitis. Participants Forty-three patients with severe acute pancreatitis, 12 patients with pancreatic cancer and 28 non-cancer controls undergoing laparoscopic cholecystectomy. Methods Plasma DNA was purified and quantified using the RNase P transcription assay and quantitative PCR. In pancreatitis patients, baseline samples were taken on admission and further samples taken at a median of 5 days into the disease course. Results Plasma DNA levels on admission in patients with acute pancreatitis (median: 0.40 ng/μL; range: 0.05-0.79 ng/μL) were significantly (P<0.001) lower than in noncancer controls (median: 1.60 ng/μL; range: 0.45-9.10 ng/μL). In patients with acute pancreatitis, DNA levels significantly (P<0.001) fell during the disease course to a median value of 0.08 ng/μL (range: 0-0.53 ng/μL). Conclusion This is the first study to use quantitative PCR to measure free plasma DNA in severe acute pancreatitis. The results show that plasma DNA is lower in patients with acute pancreatitis compared to control and that values fall further during the disease course
DNA; Pancreatitis, Acute Necrotizing
Current knowledge of the pathophysiology of acute pancreatitis suggests that acinar injury serves as the cellular trigger and that subsequent progression to a systemic illness involves a complex interplay between the pancreatic parenchymal microvasculature, circulating soluble cytokine mediators, cellular mediators of inflammation and regional endothelial beds especially those in the lung, liver and kidneys [1, 2, 3, 4].
Pancreatic necrosis is a characteristic feature of the more severe forms of this disease and may in part be mediated by occlusion of pancreatic microvasculature [5]. Radiologically, pancreatic necrosis is characterized by areas of low or absent pancreatic perfusion on intravenous contrastenhanced tomography [6]. The site and extent of necrosis correlate to disease severity [7]. In cancer, tumours release a substantial amount of deoxyribonucleic acid (DNA) into the systemic circulation [8, 9, 10, 11]. These double-stranded fragments carry somatic oncogene or tumour suppressor gene mutations found in cancer cells together with tumour-specific epigenetic alterations such as hypermethylated sequences, suggesting that most plasma DNA is derived from tumour cells [10]. Therefore, a number of studies have considered mutation analysis of candidate genes in plasma DNA as a surrogate for somatic variation in tumours. Notably, this approach has been applied to Kras mutations in individuals with pancreatic cancer [12]. In vitro studies in a murine liver cell model using acetaminophen to induce liver cell necrosis and staurosporine to produce apoptosis have shown that DNA is released into plasma by both necrotic and apoptotic pathways [13].
Plasma shedding of DNA has also been reported in non-cancer acute illnesses such as stroke [14]. In Rainer’s study, median plasma DNA concentrations taken within 3 hours of onset of symptoms were significantly higher in patients who died compared to survivors and the authors speculated that plasma DNA was an indicator of cellular injury [14]. To date; there is little evidence on the levels or relevance of plasma DNA in acute pancreatitis. Is DNA shed from the inflamed necrotic pancreas or does microvascular thrombosis prevent DNA from injured cells reaching the circulation? We postulated that DNA released by autolysing pancreatic tissue may reflect acinar injury and hence correlate with clinical outcome. Therefore, this prospective study investigates genomic DNA in plasma in patients with severe acute pancreatitis comparing measurements on admission and at a later stage in the disease course with control samples from patients with pancreatic cancer and individuals undergoing cholecystectomy.
Patients
Forty three patients admitted with a clinical diagnosis of acute pancreatitis on the basis of acute abdominal pain plus at least a threefold elevation of serum amylase constitute the study group. Patients were categorized on admission using the Acute Physiology and Chronic Health Evaluation (APACHE) II scoring system [15] and individuals with a score equal to, or greater than, 8 within 48 hours of admission to hospital are included and would correctly be regarded as “predicted severe acute pancreatitis”. Current Atlanta consensus terminology [16] was used for disease categorization and definition of complications. All case notes were reviewed after discharge and 14 (33%) patients with an admission APACHE II equal to or greater than 8 were re-categorised according to Atlanta consensus conference criteria [16] as mild acute pancreatitis as they had no evidence of organ dysfunction and a short, uncomplicated in-patient stay. Data from these patients are retained in the analyses. In all patients, data were collected in the following categories (Table 1): age, gender, probable aetiology, APACHE II score on admission and at 48 hours post-admission, logistic organ dysfunction score (LODS) at admission and at 48 hours, C-reactive protein at 48 hours, and overall mortality. Patients underwent computed tomography at the discretion of their attending (consultant) clinician. The radiology department utilised a standard “pancreatic” protocol comprising oral contrast, arterial and delayed venous phase images. Films were reported by attending (consultant) radiologists according to the Balthazar scoring system [6]. CT scans were done within 48 hours of the first DNA sample in 13 patients and showed evidence of necrosis in 8 patients (62%).
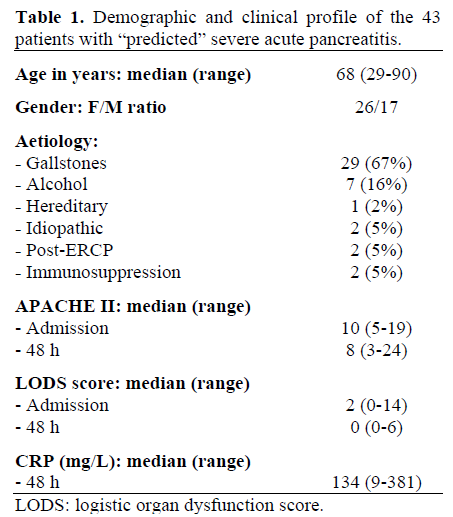
Overall, there were four deaths in this series giving a mortality of 9%. For outcome analysis, respiratory dysfunction [17, 18] was defined according to the lung injury severity criteria [19] and renal dysfunction according to the LODS [20].
The control groups comprised 12 patients with histologically-confirmed pancreatic cancer and 28 non-cancer controls undergoing elective laparoscopic cholecystectomy for chronic cholecystitis. DNA was from an anonymised databank and no demographic data were available for these two control groups.
Venous blood was drawn for analysis on the day of admission with a delayed sample being taken on approximately the 5th hospital day (median: 5 days; range: 3-7 days).
Blood samples in EDTA were centrifuged for 10 minutes at 2,000 rpm to separate plasma. Aliquots of 500 µL were stored at -80 °C prior to processing.
DNA Extraction
DNA was isolated from plasma using the QIAamp DNA micro kit (Qiagen, Valencia, CA, USA). In brief, .EDTA plasma samples were lysed under highly denaturating conditions at 56 °C in the presence of proteinase k along with buffer ATL (Qiagen, Valencia, CA, USA) followed by incubation in 100% ethanol. Optimum binding of genomic DNA in the lysate was assisted by the buffer AL (Qiagen, Valencia, CA, USA) from the kit. Carrier RNA (Boehringer Mannheim, Richfield, CT, USA) was added to buffer AL to aid binding of DNA to the elution column before precipitation. Lysates were transferred onto a QIAamp mini-e column where the DNA was absorbed onto the silica-gel membrane. Co-centrifugation of the filtrate with buffer AW1 and buffer AW2 removed contaminants. DNA was eluted with buffer AE (Qiagen, Valencia, CA, USA), incubated at room temperature for 1 minute and then centrifuged at 14,000 rpm for one minute. Initial DNA quantification was conducted using ultraviolet spectrophotometry technique (Nanodrop ND- 1000 UV, Applied Biosystems, Foster City, California, USA).
DNA Quantification
As the extraction method utilised carrier RNA to improve DNA yields, quality and concentration was quantified by human nucleic acid specific RNase P assay. RNase P gene is a single copy gene encoding the RNA moiety for the RNase P enzyme. The TaqMan RNase P detection reagents (TaqMan 7900 HTReal time Applied Bio Systems, Alameda, CA, USA) kit contains a mix of primers and probe (FAMTM dye labelled) that can be used to detect and quantify genomic copies of human RNase P gene which are designed according to Primer Express guidelines for quantitation utilizing the universal thermal cycling parameters. TaqMan Universal master mix is designed to perform a 5’ nuclease assay with the TaqMan core PCR reagents kit. A calibration curve was constructed using known concentrations of DNA and all data end points were analysed in duplicate. All procedures followed the manufacturer’s instructions.
Outcomes
The study examines the following outcomes:
• descriptive report of plasma DNA levels in severe acute pancreatitis on admission and at day 5 (median value);
• correlation between plasma DNA and outcome;
• correlation between pancreatic necrosis and plasma DNA.
Data are presented as medians and ranges. Comparisons are made by means of nonparametric analyses (Mann-Whitney U-test and Wilcoxon’s matched pairs signed ranks test). The StatsDirect version 2.4.5 software package was used (StatsDirect Ltd, Sale, Cheshire, United Kingdom; www.statsdirect.com).
The study was approved by the Local Research Ethics Committee. Written informed consent was obtained from each patient and the study protocol conforms to the ethical guidelines of the "World Medical Association Declaration of Helsinki - Ethical Principles for Medical Research Involving Human Subjects" adopted by the 18th WMA General Assembly, Helsinki, Finland, June 1964, as revised in Tokyo 2004.
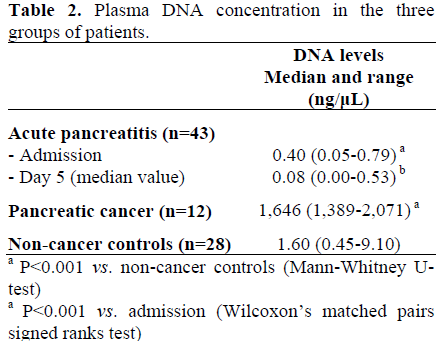
Plasma DNA Concentrations
Admission plasma DNA levels in patients with severe acute pancreatitis (Table 2) were lower than in non-cancer controls (P<0.001; Mann-Whitney U-test). In patients with acute pancreatitis, plasma DNA levels were lower at the second sampling time point at a median of day 5 compared to admission (P<0.001; Wilcoxon’s matched pairs signed ranks test). Plasma levels of DNA in patients with pancreatic cancer were grossly elevated compared to non-cancer controls (P<0.001; Mann-Whitney U-test).
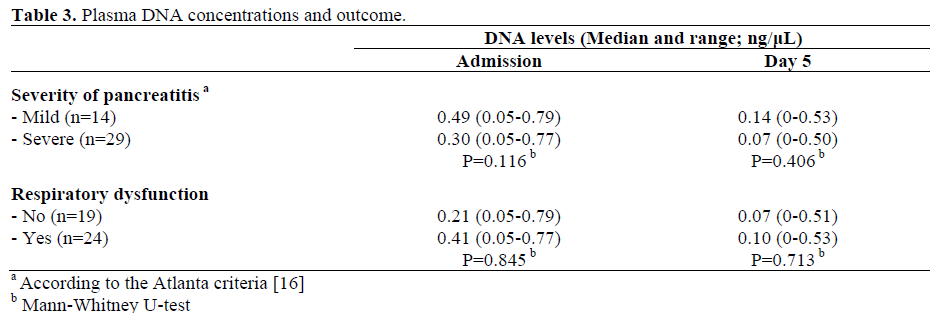
Plasma DNA Concentrations and Outcome
There was no significant difference in plasma DNA between patients with mild disease and those with eventual severe disease, as well as, plasma DNA was not significantly related to the presence of both respiratory and renal dysfunction (Table 3).
Plasma DNA Concentrations and Necrosis
No significant correlation was found between early CT evidence of necrosis and plasma DNA levels both at admission (presence of necrosis: median 0.35 ng/µL, range 0.11-0.77 ng/µL; no evidence of necrosis: median 0.58 ng/µL, range 0.10-0.68 ng/µL; P=0.833) and at day 5 (presence of necrosis: median 0.20 ng/µL, range 0.03-0.50 ng/µL; no evidence of necrosis: median 0.33 ng/µL, range 0-0.46 ng/µL; P=0.833).
This study undertakes a quantitative analysis of plasma DNA in human acute pancreatitis. The inclusion criteria aimed to identify individuals with severe disease as it was thought that these patients would be more likely to have evidence of pancreatic cellular disruption and necrosis than those with the mild form. The first issue is whether the patients in this study did indeed have severe disease as an APACHE II score equal to, or greater than, 8 within 48 hours of admission to hospital will predict 88% of severe episodes [16]. The principal inclusion criterion for this study was thus set as an APACHE II score equal to, or greater than, 8. However, accepting that not all patients with an APACHE II score equal to, or greater than, 8 will have severe disease, other indicators of severity include evidence of necrosis (62%, 8 of 13 patients undergoing CT done within 48 hours of the first DNA sample) and overall mortality (9%). It seems reasonable to regard this cohort as representative of severe acute pancreatitis but to consider that different results may be obtained in patient groups with more severe disease.
The first key finding is that plasma DNA in severe acute pancreatitis is lower than in controls and that these values decrease at the second sampling time-point. Technical error in analysis or quantification seems unlikely as an explanation as the high values obtained in the “positive” control group of patients with pancreatic cancer are similar to the values obtained by other groups [13]. Digestion of released DNA by activated proteases is a possibility that must be considered. Although this study did not seek to quantify base pair lengths, short fragments of 100 bp would have been detected by the methods employed. The results of this study provide the first in vivo correlate for Dembinski’s findings [21]: in an ischaemia-reperfusion rat model of acute pancreatitis they found that DNA synthesis was maximally reduced (by 70%) compared to control within 24 hours of onset of the illness with gradual recovery over the ensuing four weeks
Reflecting the greater complexity of the human illness, no significant correlation was observed between CT evidence of necrosis and plasma DNA. However, as only a small number of patients in this study underwent CT close to the timepoint of blood sampling for DNA this issue requires further investigation.
In summary, this study is the first to quantitate plasma DNA in human severe acute pancreatitis. In contrast to other acute severe illnesses including cerebrovascular accident and studies of patients requiring intensive care treatment, plasma DNA levels are not raised. This is thus an important negative finding. Furthermore, in contrast to pancreatic cancer, plasma DNA from patients with acute pancreatitis does not provide a good source of material to explore potential somatic gene changes important in the pathogenesis of the disease. Further studies are required to establish whether the fall in DNA seen in our pancreatitis group represents a genuine host response to pancreatic injury, or whether necrotic or apoptotic pathways predominate and whether DNA levels are restored to normal in those individuals recovering from this illness.
William Newman was supported by a grant from the Royal Society