Original Research Article - (2022) Volume 7, Issue 6
Reaching the Remote: Dried Blood Spot Analysis for Disease Diagnosis on a Protein Microarray Platform
Metoboroghene O. Mowoe*,
Tristan Rensburg,
Bradley Africa,
Eduard Jonas and
Jonathan M. Blackburn
1Department of Integrative Biomedical Sciences, University of Cape Town, South Africa
2Department of General Surgery, University of Cape Town, South Africa
*Correspondence:
Metoboroghene O. Mowoe, Department of Integrative Biomedical Sciences, University of Cape Town,
South Africa,
Email:
Received: 01-Jun-2022, Manuscript No. IPJHCC-22-13720;
Editor assigned: 03-Jun-2022, Pre QC No. IPJHCC-22-13720 (PQ);
Reviewed: 17-Jun-2022, QC No. IPJHCC-22-13720;
Revised: 22-Jun-2022, Manuscript No. IPJHCC-22-13720 (R);
Published:
29-Jun-2022, DOI: 10.36846/IPJHCC-7.6.70026
Abstract
Context: Cancer remains one of the leading causes of death globally with an estimated 19.3 million cases and 10
million mortalities in 2020. In Africa and Asia, where remoteness is prevalent, access to healthcare facilities is limited,
providing a significant barrier to effective screening and early detection of cancers in at risk groups and thus,
incomplete registries.
Objectives: Here, we utilised low resource, low cost dried blood spots (DBS) based sample collection coupled with
robust, protein microarray technology to enable quantitative, multiplexed measurements of diagnostic auto antibody
biomarkers of disease, in minimal sample volumes. I
Methods: Specifically, we describe the development of a DBS extraction and elution method from low cost, homemade
blood cards. We then show that DBS stored at room temperature (25℃, RT) for up to 15 d yield comparable
autoantibody signatures to autologous serum samples stored at -80℃ and those from samples prepared on a
commercially available blood card. We further conducted a pilot study, comparing total IgG and three previously
identified autoantibodies up regulated in pancreatic cancer (PC), in DBS from 11 PC patients stored at RT for up to
15 d.
Results: We found comparable protein profiles across commercially developed blood cards and our low cost, in
house kit with no significant difference in autoantibody profiles over 15 d (p>0.05).
Conclusion: Such low cost, DBS based sample collection methods, combined with regular, RT courier shipments
and ultrasensitive protein microarray based detection in a remote laboratory, thus have the potential to facilitate
future, unbiased, large scale serosurveys and serological diagnostic testing within remote, rural communities.
Keywords
Dried blood spots; Diagnosis; Cancer; Remote; Communities; Rural populations
Introduction
Geographic variations in cancer incidence, and consequentially
survival, are partly due inequity and inequality in terms of global
access to healthcare and the quality of registries in remote,
rural populations [1,2]. Thus, reported disease incidence rates
are often inversely correlated to rural population proportions
in different regions (Figure 1), despite a worse health status
in remote, rural areas than urban areas [3,4]. This implies dramatic
under reporting in such areas due to limited access to screening and is most prominent in middle and low income
countries (LMICs), most of which exist on the African and Asian
continents.
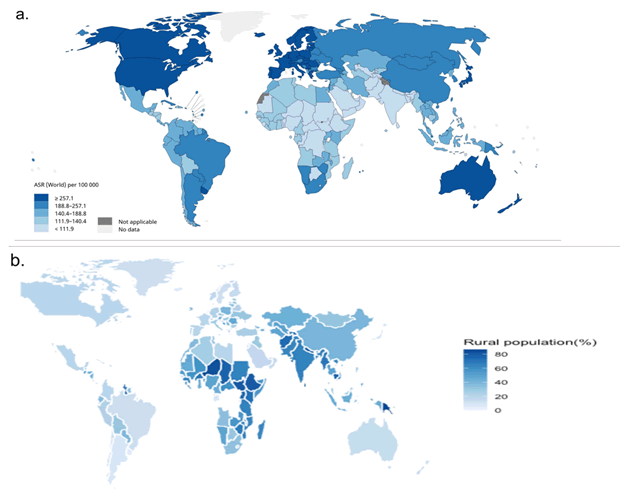
Figure 1: Maps of global cancer incidence and rural population percentage a.) Estimated age-standardized global incidence rates for all cancers, including both sexes and ages in 2020 (produced in http://gco.iarc.fr/today/home) [22], b.) Percentage of global rural population (data derived from https://data.worldbank.org/indicator/SP.RUR.TOTL.ZS?end=2019&start=1960&view=map) and plotted using ggplot [26] in R.
Limited access to diagnostic tools, funding for diagnostic and
laboratory services and specialists, combined with patient related
barriers to follow through with diagnostic referrals, impede
disease diagnosis in these communities [5]. To overcome
diagnostic limitations, dried blood spots (DBS) have been used
as an analytical matrix in the clinical setting for over half a century,
primarily for disease screening of neonates [6-8]. They are minimally invasive, require little to no training and storage infrastructure,
and obviate the risk associated with needles and
syringes used in venous blood collection. However, the minimal
recovered sample volumes can create technical challenges for
downstream protein biomarker measurements that lack the
massive signal amplification of PCR methods.
In cancers, autoantibodies (Aabs) have gained recent popularity
as candidate biomarkers since they exhibit increased levels
during the early stages of disease and can potentially predict
disease progression and treatment outcomes [9]. Increases in
Aab concentration may be detectable months to years before
clinical symptom presentation [10-13] and other biomarkers
are measurable, making them ideal diagnostic biomarker candidates.
However, it is increasingly clear that multiplexed panels
of Aab biomarkers are required to provide clinically useful
early diagnostic performance [14], which in turn poses technological
challenges in multiplexing classic ELISA measurements,
both in terms of sample volumes required and assay costs.
Protein microarrays, in principle, can solve these problems by
providing a miniaturised, highly multiplexed, ultra-sensitive
ELISA-like assay format and have been used, successfully, to
monitor disease activity [15] and discover novel autoantibody
biomarkers [16] in serum. Moreover, previous studies have
shown that therapeutic antibody titres measured in DBS are
comparable to those measured in serum and plasma [17,18],
suggesting that the combination of DBS based sample collection
with protein microarray based autoantibody detection
might enable screening of at risk remote populations for early
cancer biomarkers. Here, we describe the development and
validation of a robust protein microarray based method for the
quantitation of autoantibody profiles in DBS from patients with
chronic pancreatitis (CP) and pancreatic cancer (PC).
Materials and Methods
Sample Collection and Dried Blood Spots
Inclusion criteria for method development and validation were
blood samples from one CP patient prior to resective surgery
and 11 randomly selected, late stage PC patients, respectively
(Table 1). This study was approved by the University of CapeTown
Human Research Ethics Committee (HREC 559-2018).
Written informed consent was obtained from individuals for
whom study samples were derived.
Table 1: Demographic and Clinical Characteristics of Patients from which Dried Blood Spots were extracted.
| Variable |
Disease cohort |
|
Chronic pancreatitis (n=1) |
Pancreatic cancer (n=11) |
| Age (y) |
49.0 ± 5.00 |
61.82 ± 11.37 |
| Sex (n) |
| Male |
1 |
4 |
| Female |
- |
7 |
| Race (n) |
| Black |
1 |
4 |
| Coloured |
- |
5 |
| Other |
- |
2 |
In-house DBS cards: Here, 50 μL of whole blood from all 12
patients was pipetted unto Whatman™ filter paper 1 (150 mm;
Cat#1001150) and allowed to dry for 1 h, at room temperature
(23, RT). The filter paper was then placed in resealable plastic
bags with a drierite desiccant (#737828-454G, Sigma-Aldrich)
and stored in the dark, until ready for further use.
Commercial DBS cards: ArrayIT blood cards were used to compare
and validate the results from our low cost, homemade
cards. Following collection, 50 μL of whole blood from the CP
patient was pipetted unto ArrayIT dried blood cards (#ABC, ArrayIT
Corporation) and allowed to separate into red blood cell
components and serum for 5 min. The blood cards were then
left to dry at RT for 5 min, according to the manufacturers’ instructions,
placed in the anti-static shipping envelopes provided,
and stored in the dark, until ready for further use.
Serum control: Following DBS collection, whole blood was centrifuged
at 1300 × g for 13 mins, and serum was isolated and
stored at -80℃ until ready for further use.
Sample Extraction
In house Whatman™ DBS cards: Based on previous DBS methods
and the requirements for microarray assays, a method
of serum extraction from DBS was developed. On days 1, 5,
10, and 15, a 5 mm disc was excised from blood spots on the
Whatman™ filter paper using a disposable punch (#MT3336,
Integra Miltex). The filter discs were handled with fine tipped
forceps to prevent contamination. Each disc was soaked in 250
μL Phosphate Buffered Saline containing 0.1% Tween 20 (PBST)
in a 24 well plate and incubated at RT on a shaker for 60 min
× 100 rpm. Subsequently, the eluent was placed in 1.5 mL microfuge
tubes and centrifuged at 14,000 g × 10 min to sediment
cell debris, after which each supernatant was transferred to a clean tube.
Commercial ArrayIT DBS cards: To validate our in house Whatman™ filter paper method, we extracted serum from ArrayIT
blood cards based on the manufacturers’ instructions on days
1, 5, 10, and 15. A 5 mm disc was excised from the serum portion
of the blood card, presumed to contain antibodies, antigens,
and other serum proteins. The disc was wet with 10 μL of
PBST, placed in 1.5 mL microfuge tubes, and allowed to rehydrate
for 30 mins at RT. Subsequently, the disc was centrifuged
at 14,000 g × 1 min to elute the serum into the collection tube.
The disc was then rewet with 10 μL of PBST and re-centrifuged
to elute any remaining bound proteins. The eluents were combined,
bringing the final volume to 20 μL.
Microarray Analysis
To determine the optimal dilution factor of the DBS eluents
from each card for the assays, a microarray assay including 5
control spots of known concentrations was performed (Figure 2). We found that, similar to
the serum control, a 1:800 dilution of ArrayIT eluent produced
an optimal.
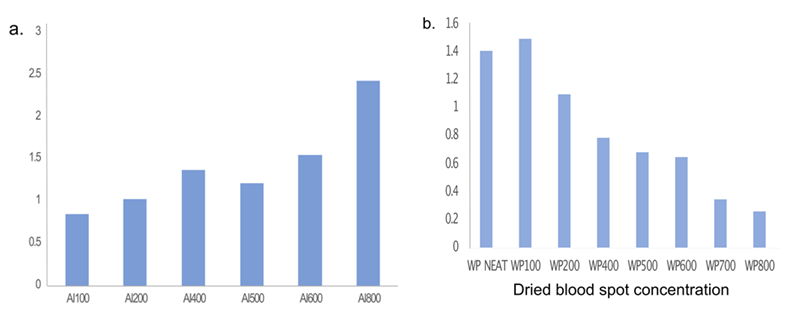
Figure 2: Protein expression to determine optimal dilution for a.) ArrayIT and b.) Whatmann dried blood spot eluents for subsequent microarray assays. *Ai(n) – ArrayIT dried blood card (dilution factor); W ()– Whatmann filter paper dried blood card (dilution factor).
Method development: For the microarray assays, Sengenics
IMMUNOMETM arrays pre-printed with 1622 proteins on each
array were used. Prior to the assays, the slides were removed
from their storage solution and washed 3×5 min in PBST and
1×5 min in ddH2O. The serum and eluents from each method
were diluted based on the dot blot assay and slides were incubated
with sample on an orbital shaker at 100 rpm for 60 min in
a light protected slide processing dish to prevent photobleaching.
Subsequently, slides were washed 3×5 min and rinsed 1×5 min in PBST and ddH20, respectively. Slides were then incubated
in 20 μg/ml of detection antibody (Alexa-fluor 647 coupled
goat anti-human IgG (H+L); Cat#A21445, ThermoFischer
Scientific) for 30 min on an orbital shaker at 100 rpm for 30
min. Again, slides were washed in PBST and ddH20, respectively,
and then dried via centrifugation at 1300 RCF × 3 min at RT.
Dried slides were scanned according to pre-set parameters and
saved as TIFF files, which were used for data extraction downstream.
Validation: The assay process was replicated on 11 PC patients
using a custom made chip including 3 proteins that had previously
been identified (unpublished data) to be upregulated in
PC patients (MAGEA5, MART.1, NY.CO.45) and a total anti-human
IgG control.
Statistical Analysis
The microarray image data was extracted using Mapix (v 8.5.0) and the Sengenics IMMUNOMETM gal file, and median foreground
and background intensities were read into RStudio
(v.4.0) for pre-processing using the Pro-MAP single channel microarray
analysis pipeline script (developed by MOM). Briefly,
non-specific binding, array data that was not significantly different
to surrounding background (defined as spot intensities
<2SD of the median background) were filtered out. Data was
then normexp background corrected and cyclic loess normalized
prior to downstream analysis. A one way analysis of variance
(ANOVA) was used to compare the average log expression
intensity of proteins from the three sample collection methods
(serum, ArrayIT, and Whatman™ DBS). Subsequently, a
limma linear model using calculated array weights, was fit to
the normalized microarray data to fully model the systematic
part of the data and determine variability between the groups
using the limma package in R [19]. To determine variability in
the data based on the comparisons of interest, we extracted
contrasts matrices. In this way, we were able to determine if
there were differences between: 1. eluents from ArrayIT and
Whatman™ DBS methods, and 2. eluents from the two DBS
methods on Day 1, 5, 10, and 15. Subsequently, an empirical
Bayes method was used to moderate the standard errors of the
estimated log fold changes.
Finding no differences between methods or across days we
then compared Aab profiles of our Whatman™ method over
15 d. To determine variability in Aab profiles of the 11 PC patients
over 15 d for validation of our findings, a limma time
course analysis was run. The probability level of p<0.01 was
determined as differentially expressed. All statistical analyses
were conducted using R (version 3.6.0) and all result images
were created using the ggplot2 package in R [20]. Power analysis
for an effect size=0.8, yielded a power=0.805.
Results
Comparison of Serum Control to Dried Blood
Ccard Methods
Median log expression intensity of proteins from the CP serum
control (9.67 ± 0.745) was slightly higher than that of the corresponding
ArrayIT (9.60 ± 0.834) and Whatman™ (9.63 ± 0.675)
eluents. However, there was no significant difference between
the average log expression of the serum control versus Array-
IT (t=-2.820, and p=0.013) and Whatman™ eluents (t=-1.421,
p=0.330) (F=3.977, p>0.01) (Figure 3a). Furthermore, the MA
plot of the protein profiles from all three samples on day 1
showed normalized log fold change values (M) close to zero
(Figure 3b). A density plot of the serum control and two DBS
methods showed a shift of the peak density to lower average
log-expression values for the two DBS methods (Figure 3c),
implying that the DBS samples had lower non-specific background
signals.
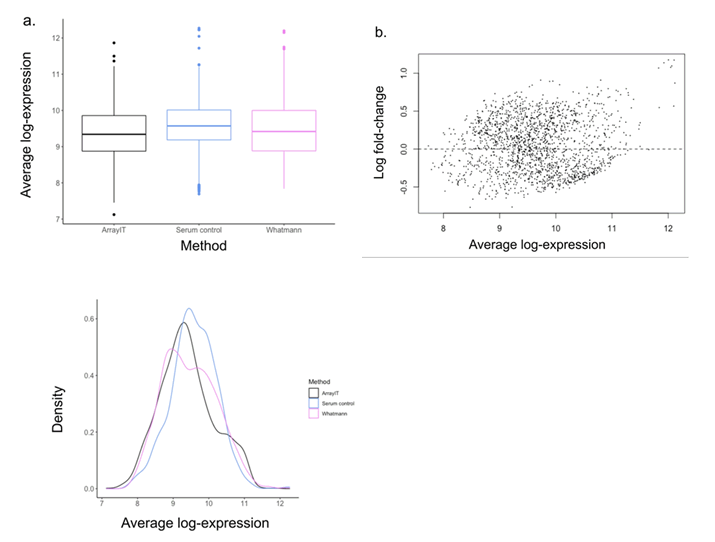
Figure 3: Comparison of average log protein expression from chronic pancreatitis serum control, ArrayIT and Whatman™ eluent samples assayed on the immunome array. a.) Boxplot comparison median average log intensities found in serum sample and ArrayIT and Whatman™ eluent samples b.) MA plot obtained comparing the three extraction methods. M represents the expression intensity of serum versus the average of the two other samples c.) Smoothed empirical densities for average protein log intensities in serum control and DBS arrays.
Comparison of Protein Expression of ArrayIT
and Whatman™ Eluent Arrays and Over Time
There was no significant difference between immunome protein
expression of eluents from the ArrayIT and Whatman™
arrays (adj. P>0.248) over all time points measured (Figures 4a
and 4b).
Concurrently, MA plot showed M values close to zero, but
slightly higher than the comparison of serum control to both
DBS eluents (Figure 4b). Furthermore, we found that there was
a slight but non-significant decrease in average log expression
over time (adj. P>0.721) (Figure 4c). Furthermore, there was a
slight decline in the abundance of protein expression over time
(Figure 4d).
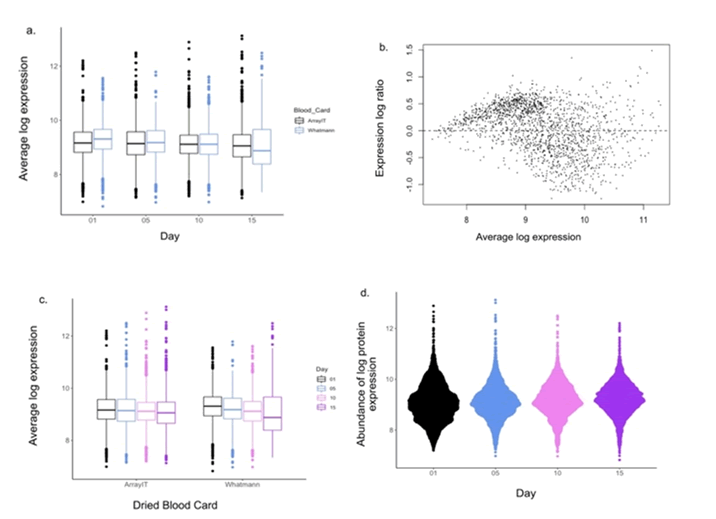
Figure 4: Comparison of average log protein expression of the two dried blood spot eluents from CP patient assayed on the immunome arrays, over time. a.) Boxplot comparing average log expression of ArrayIT and Whatman™ DBS protein arrays b.) MA plot obtained comparing the two DBS methods c.) Boxplot comparing protein expression over time in both DBS array types d.) Bee swarm plot visualizing distribution of relative maintained signal for the 1622 proteins analysed compared over 15 d.
Validation of Whatman™ DBS Method on Custom
Microarray
Multiple linear regressions: We ran a multiple linear regression
analysis to determine if any factors other than time had
an effect on protein expression intensity in the 11 PC patients
assayed on the custom DBS array. We found no effects of age,
race, or gender on protein intensity (F=0.962, p=0.493).
Time course analysis: A limma time course analysis was run to
compare protein expression intensities of the 3 proteins and
IgG controls assayed over 15 d in 11 PC patients. We found no
proteins differentially expressed in any of the time comparisons
(Figure 5a). Furthermore, we found no significant difference in
expression intensity of any of the proteins over 15 d (p<0.01)
(Figure 5b). However, as previously observed we found a reduction,
albeit non-significant, in protein abundance over time
(Figure 5c).
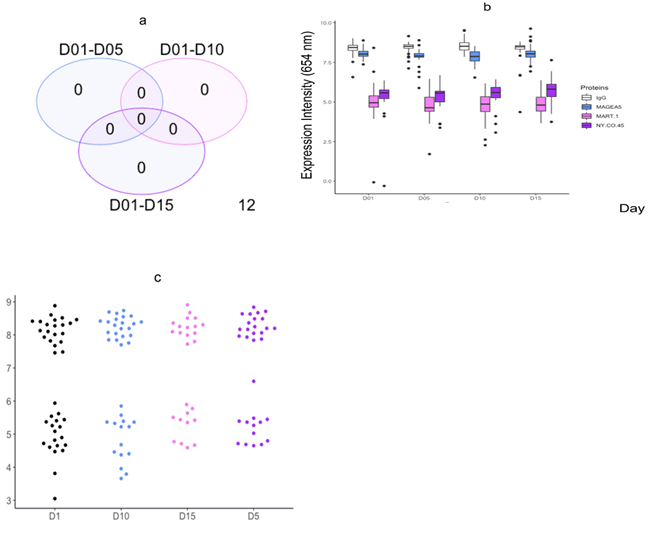
Figure 5: Comparison of protein intensity expression and abundance from day 0-15 in 11 pancreatic cancer patients assayed on the custom DBS array. a.) Venn diagram showing number of proteins significantly different in each of the comparisons of days eluted, b.) Boxplot comparing expression intensity of each of the 4 proteins over 15 d, c.) Bee swarm plot visualizing distribution of relative maintained signal for the 4 proteins analysed compared over 15 d.
Discussion
Cancer is an enormous global burden that is predicted to increase
with the growth and ageing of populations [21,22].
Unfortunately, economically disadvantaged countries face a
disproportionally high burden of infection related cancers compared
to their developed counterparts. Fortunately, the cancer
burden can be largely mitigated by early disease detection [23].
Serological testing plays an important role in early screening
and diagnosis in several disease areas and the identification
of more effective biomarkers for early detection. Serum autoantibody
profiles can indicate the presence of diseased cells
months to years prior to symptom presentation [10-13]. This
is especially useful in rare malignancies, such as PC, known to
clinically present at an advanced stage in most patients. Large
serosurveys may be key to discovering early detection tools,
especially in populations with large rural, remote communities,
where access to the associated technologies remains limiting.
This study was performed to quantitatively compare results derived
from serum obtained from venous blood collection under
routine conditions and blood dried on two different DBS cards,
commercial (Array IT) and homemade (Whatman™ no 1). Several
studies have shown a correlation with data from venous
blood samples and blood dried on Whatman™ 903 substrates
[24]. However, Whatman™ no 1 differs from 903 in that the
latter is made specifically for protein retention, but the former
is more easily accessible largely due to cost and usefulness for
various other laboratory experiments. Our use of the latter
was influenced by the need for a more cost effective method of blood collection for primarily low income communities. We
developed a method of extracting eluent sample from blood
cards made using Whatman™ no 1, eliminating the need for
phlebotomies and reducing the sample collection burden on
stressed healthcare systems, especially in LMICs.
The results presented here show that eluent extracted from
these blood cards up to 15 d following blood collection yielded
results comparable to those from serum isolated from whole
blood following venous blood collection. In addition, Whatman™ eluents yielded profiles comparable to those of Array
IT blood cards, thus representing a low cost alternative to currently
available commercial blood cards which typically cost
~USD10/sample. Notably, protein expression intensities from
the serum control sample remained higher, albeit non-significantly,
compared to DBS eluents, but simultaneously, the DBS
samples appeared to present with a lower background, perhaps
due to permanent absorption of larger macromolecules
and complement factors on to the membranes.
The dried blood spot modality has the potential for wide scale
screening of diseases such as cancers and autoimmune, infectious,
and cardiovascular diseases where this is necessary
to drastically reduce fatalities. The widespread use of DBS in
the past has been impeded by small sample volumes and low
target analyte concentrations requiring a sensitive and specific
assay for detection and quantification. The use of protein
microarray methods for DBS eluent testing has the unique advantage
of requiring minimal volumes (1 μl) per assay, without
compromising the ultra-sensitivity (pg/ml detection range) of
the resultant assays [25]. Thus, DBS samples can be used for
multiple testing or to support further analyses. Future studies
will investigate the feasibility of a self-collection kit for at home
finger prick DBS collection using the method developed here to
elute and test samples that show comparable analytical performance
to venepuncture derived blood samples [26].
Conclusion
Overall, this study could pave the way to large scale serosurveys
in remote populations by utilizing simple, low cost DBS sample
collection, combined with RT courier shipments to a centralized
testing laboratory and miniaturized, protein microarray
based quantitative, multiplexed biomarker detection, thereby
increasing the effectiveness of screening campaigns and early
diagnosis, and the accuracy of global cancer registries.
Author Contributions
Conceptualization: JMB, MOM; Methodology, Validation, Formal
analysis, and Investigation: MOM; Resources: MOM, JMB,
EJ, UK; Data curation: MOM, TR, HA, JG, UK, MB, BA, EJ; Writings
Original Draft: MOM; Writing Review and Editing: MOM, JMB,
EJ; Visualization: MOM; Supervision: JMB, EJ; Project administration:
MOM; Funding acquisition: JMB, EJ. The manuscript
was written through contributions of all authors. All authors
have given approval to the final version of the manuscript.
REFERENCES
- Valsecchi MG, Steliarova-Foucher E (2008) Cancer registration in developing countries: Luxury or necessity?. Lancet Oncol. 9(2):159-167.
[Crossref] [Google Scholar] [PubMed]
- Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA: A Cancer J Clinicians. 55(2):74-108.
[Crossref] [Google Scholar] [PubMed]
- Strasser R (2003) Rural health around the world: Challenges and solutions*. Fam Prac. 20(4):457-463.
[Crossref] [Google Scholar] [PubMed]
- Krümmel EM (2009) The circumpolar inuit health summit: A summary. Int J Circumpolar Health. 68(5):509-518.
[Crossref] [Google Scholar] [PubMed]
- Anticona Huaynate CF, Monica JPT, Malena C, Holger MM, Richard O, et al. (2015) Diagnostics barriers and innovations in rural areas: Insights from junior medical doctors on the frontlines of rural care in Peru. BMC Health Serv Res. 15(1):454.
[Crossref] [Google Scholar] [PubMed]
- Guthrie R, Susi A (1963) A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatr. 32(3):338.
[Crossref] [Google Scholar] [PubMed]
- Eyles DW, Ruth M, Cameron A, Pauline K, Thomas B, et al. (2010) The utility of neonatal dried blood spots for the assessment of neonatal vitamin D status. Pediatr Perinatal Epidemiol. 24(3):303-308.
[Crossref] [Google Scholar] [PubMed]
- Crossley JR, Smith PA, Edgar BW, Gluckman PD, Elliott RB (1981) Neonatal screening for cystic fibrosis, using immunoreactive trypsin assay in dried blood spots. Clinica Chimica Acta. 113(2):111-121.
[Crossref] [Google Scholar] [PubMed]
- Patel AJ, Tan TM, Richter AG, Naidu B, Blackburn JM, et al. (2022) A highly predictive autoantibody-based biomarker panel for prognosis in early-stage NSCLC with potential therapeutic implications. Br J Cancer. 126(2):238-246.
[Crossref] [Google Scholar] [PubMed]
- Anderson KS, LaBaer J (2005) The sentinel within: Exploiting the immune system for cancer biomarkers. J Proteome Res. 4(4):1123-1133.
[Crossref] [Google Scholar] [PubMed]
- Lernmark A (2001) Series Introduction: Autoimmune diseases: Are markers ready for prediction?. J Clin Invest. 108(8):1091-1096.
[Crossref] [Google Scholar] [PubMed]
- Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Gregory JD, et al. (2003) Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 349(16): 1526-1533.
[Crossref] [Google Scholar] [PubMed]
- Henriksson G (2014) Presymptomatic autoantibodies in Sjögren’s syndrome: What significance do they hold for the clinic?. Exp Rev of Clin Immunol. 10(7):815-817.
[CrossRef] [Google Scholar] [PubMed]
- Zaenker P, Ziman MR (2013) Serologic autoantibodies as diagnostic cancer biomarkers:A review. Cancer Epidemiol Biomarkers Prev. 22(12): 2161-81.
[Crossref] [Google Scholar] [PubMed]
- Robinson WH, Fontoura P, Lee BJ, Neuman de Vegvar HE, Tom J, et al. (2003) Protein microarrays guide tolerizing DNA vaccine treatment of autoimmune encephalomyelitis . Nat Biotechnol. 21(9):1033-1039.
[Crossref] [Google Scholar]
- Price JV et al. (2013) Protein microarray analysis reveals BAFF-binding autoantibodies in systemic lupus erythematosus. J Clin Invest. 123(12):5135-45.
[Crossref] [Google Scholar] [PubMed]
- Prince PJ, Matsuda KC, Retter M, Scott G (2010) Assessment of DBS technology for the detection of therapeutic antibodies. Bioanal. 2(8):1449-1460.
[Crossref] [Google Scholar] [PubMed]
- Kaendler K, Warren A, Lloyd P, Sims J, Sickert D (2013) Evaluation of dried blood spots for the quantification of therapeutic monoclonal antibodies and detection of anti-drug antibodies. Bioanal. 5(5):613-622.
[Crossref] [Google Scholar] [PubMed]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, et al. (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7):e47.
[Crossref] [Google Scholar] [PubMed]
- Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York.
[Google Scholar]
- Omran AR (2005) The epidemiologic transition: A theory of the epidemiology of population change.1971. Milbank Q. 83(4):731-757.
[Crossref] [Google Scholar] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, et al. (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians.
[Crossref] [Google Scholar] [PubMed]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, et al. (2015) Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 65(2):87-108.
[Crossref] [Google Scholar] [PubMed]
- Moretti M, Freni F, Tomaciello I, Vignali C, Groppi A, et al. (2019) Determination of benzodiazepines in blood and in dried blood spots collected from post-mortem samples and evaluation of the stability over a three-month period. Drug Test Anal. 11(9):1403-1411.
[Crossref] [Google Scholar] [PubMed]
- Beeton-Kempen N, Duarte J, Shoko A, Serufuri JM, John T, et al. (2014) Development of a novel, quantitative protein microarray platform for the multiplexed serological analysis of autoantibodies to cancer-testis antigens. Int J Cancer. 135(8):1842-1851.
[Crossref] [Google Scholar] [PubMed]
- Hadley W (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016.
[Google Scholar]
Citation: Mowoe MO, Rensburg T, Ali H, Gqada J, Kotze U, et al. (2022) Reaching the Remote: Dried Blood Spot Analysis for Disease
Diagnosis on a Protein Microarray Plaform. J Healthc Commun. 7:70026
Copyright: © Mowoe MO, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution
License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source
are credited.