Research Article - (2022) Volume 12, Issue 6
Received: 08-Jun-2020, Manuscript No. EJEBAU-21-4565; Editor assigned: 11-Jun-2020, Pre QC No. EJEBAU-21-4565 (PQ); Reviewed: 25-Jun-2020, QC No. EJEBAU-21-4565; Revised: 03-Aug-2022, Manuscript No. EJEBAU-21-4565 (R); Published: 31-Aug-2022, DOI: 10.36648/2248-9215.12.5.146
Interleukin 17-A (IL17A) is one of the major cytokines known to play a role in different pathologies, including Autoim- mune Diseases (AIDs). IL17A gene has different Single Nucleotide Polymorphisms (SNPs); rs2275913 being frequent- ly studied SNP with susceptibility to autoimmune diseases. However, the results for this SNP have remained incon- clusive. Therefore, in this meta-analysis we gathered data from different studies conducted in different populations (Europe, Asia, New Zealand, Africa and Latin America) to find out the role of rs2275913 in overall susceptibility of AIDs. After thorough search and careful screening, a total of 5424 cases and 7221 controls were cumulatively found in 16 eligible studies from 13 different research papers. Analyses were done under co-dominant homozygous (AA vs. GG), co-dominant heterozygous (GA vs. GG), dominant (AA+GA vs. GG), recessive (AA vs. GG+GA) and allelic (A vs. G) models. All the eligible studies were in line with Hardy-Weinberg equilibrium and the I2 values of all models were <50%. Significant Q values were found only for the dominant (p=0.09) and allelic (p=0.07) models. No publication bias was seen when funnel plots were constructed. Significant protective effect of an allele (OR<1, p<0.05) was ob- served in some individual studies on rheumatoid arthritis and periodontitis. However, the results of meta-analysis in total fixed effect and total random effect of all the studies in different models showed no significant association. As per the best of our knowledge, this is the first report of meta-analysis of rs2275913 of IL17A gene with respect to autoimmune diseases susceptibility.
Autoimmune disease; Interleukin-17A; Rheumatoid arthritis and periodontitis
Autoimmune Diseases (AIDs) are caused by specific immune responses directed against body’s own structures due to the failure of the self-tolerance [1]. There are different tolerance mechanisms in a healthy immune system to protect against the activation of self-reactive lymphocytes. In autoimmune diseases, there is a failure of one or more of these tolerance mechanisms leading to activation and persistence of auto-reactive T and B cells [2]. In addition to loss of tolerance, there are other factors that characterize autoimmune diseases including imbalance in the pro-and anti-inflammatory processes and chronic inflammation resulting from increased levels of inflammatory and mediatory cells and autoantibodies [3]. Although AIDs are diverse and affect multiple organs, there is a remarkable similarity in the underlying mechanisms [4].
Interleukin 17 (IL17) is a group of cytokines released by Th17 cells, a type of T helper cells. The consistent release of IL17 from Th17 cells favors chronic inflammation [5,6]. Hence, IL17 has been implicated in the pathogenesis of autoimmune diseases such as Rheumatoid Arthritis (RA), Systemic Lupus Erythematosus (SLE), Lupus Nephritis (LN) and Sjogren’s Syn- drome (SS) [8-10].
IL17 family has six members (IL17A-F), IL17A being the most prominent member playing important role in different inflammatory and autoimmune diseases [11,12]. The rs2275913 Single Nucleotide Polymorphism (SNP) of IL17A gene has been studied for association with susceptibility to different AIDs with inconsistent results [13].
Therefore owing to the involvement of IL17A cytokine in different autoimmune diseases and inconclusive results from different case-control studies, we conducted a meta-analysis of rs2275913 in different AIDs [14].
Publication Search
PubMed, Google-Scholar, Science Direct and Web of Science were used as searching tools for all the relevant articles with the following key words: “Role of IL-17A SNPs in autoimmune diseases” or “IL-17A SNPs in autoimmune disorders” or “IL-17A polymorphisms in autoimmune disorders” and “association of IL-17A with autoimmune disorders” [Higgins JPT and Green S, 2011]. We considered studies published between 2002 and 2018. Studies were checked and screened for their respective references for additional eligible studies; last search was 30th November, 2018 [15].
Selection Criteria
The inclusion criteria were defined as follows:
• Original articles
• Articles written in English
• Contained detailed genotyping information for estimating Odd Ratio (OR) and the 95% Confidence Interval (CI)
• Involved case-control designed experiment and
• Those with the genotype frequency distribution in control group observed in line with Hardy-Weinberg Equilibrium (HWE).
Exclusion criteria included:
• Unpublished articles, review articles and abstracts
• Duplication of some previous study and
• Other SNPs of the same gene [16].
Data Extraction
Two investigators jointly identified the following information from the articles: The first author, year of publication, type of the autoimmune disease, study population, number of genotyped cases and controls, genotyping method and results with respect to association of the IL17A SNP rs2275913 [17].
Statistical Methods
Hardy-Weinberg equilibrium of genotype frequencies in control group was estimated for each study through goodness-of- fit chi-square test [18]. Studies in which control group genotype frequency distribution deviated from HWE were excluded from analysis [19].
ORs and 95% CIs were calculated to find out the association of rs2275913 with susceptibility of AIDs. Random effect model was used because of varying ethnicities [20]. Cochrane’s Q homogeneity test and inconsistency (I2) index were used for the heterogeneity analysis. For Cochrane’s Q test, a p-value of less than 0.10 was considered as significant heterogeneity [21]. The I2 value of 25% was used to correspond to low heterogeneity, 50% to moderate and 75% to high degree of heterogeneity. Publication bias was assessed through funnel plots. Meta-analysis of eligible studies was carried out using MedCalc version 18.5 [22].
Study Characteristics
We found a total of 4889 relevant publications when all databases were searched during the initial screening. Among them, 160 were chosen for full text review and 4729 were excluded based on the initial exclusion criteria. Further examination was made for critical screening. We excluded 147 that did not satisfy the inclusion criteria. A total of 13 articles were selected for final analysis, three of which included two types of autoimmune diseases each, making a total of 16 studies for meta-analysis (Figure 1). A total of 5424 cases and 7221 controls for different AIDs were analyzed. Specific AIDs included rheumatoid arthritis (n=6), rheumatoid arthritis with Sjogren’s syndrome (n=1), primary antiphospholipid syndrome (n=1), periodontitis (n=1), pediatric systemic lupus erythematosus (n=1), panuveitis (n=1), myasthenia gravis (n=1), Graves’ disease (n=1), Vogt-Koyanagi-Harada syndrome (n=1), Behcet’s disease (n=1) and multiple sclerosis (n=1). Characteristics of the included studies are summarized [23].

Figure 1: Data extraction for meta-analysis of IL17A rs2275913.
Results of Meta-Analysis
Results of meta-analysis for rs2275913 are shown in Tables 1-6. Total ORs showed non-significant results in all models: Co-dominant homozygous (OR=0.97, 95% CI=0.83-1.14), co-dominant heterozygous (OR=0.99, 95% CI=0.9-1.11), dominant (OR=0.99, 95% CI=0.9-1.10), recessive (OR=0.98, 95% CI=0.87-1.1) and al lelic (OR=0.98, 95% CI=0.91-1.06) [24].
| Study first author | Disease | Ethnicity | Cases/ controls | Genotyping assay | Cases GG/GA/AA | Controls GG/GA/AA | Source of DNA |
|---|---|---|---|---|---|---|---|
| Bogunia-Kubik (2015) | Rheumatoid arthritis | Polish | 88/125 | LightSNiP | 12/44/32 | 20/67/38 | Blood |
| Pawlik (2016) | Rheumatoid arthritis | Caucasians | 417/337 | Taqman | 173/193/51 | 118/169/50 | Blood |
| Nordang (2009) | Rheumatoid arthritis | New Zealand | 580/504 | Taqman | 246/251/83 | 208/238/58 | Blood |
| Nordang (2009) | Rheumatoid arthritis | Norway | 938/920 | Taqman | 396/428/114 | 335/461/124 | Blood |
| Carvalho (2016) | Rheumatoid arthritis/Sjogren's syndrome | Brazil | 31/75 | Taqman | 37243 | 49/20/6 | Saliva |
| Carvalho (2016) | Rheumatoid arthritis | Brazil | 100/75 | Taqman | 56/38/6 | 49/20/6 | Saliva |
| Popovic-Kuzmanovic (2013) | Primary antiphospholipid syndrome | Serbia/Helsinki | 50/50 | Taqman | 17/26/7 | 18/26/6 | Blood |
| Saraiva (2013) | Periodontitis | Brazil | 116/62 | Taqman | 75/39/2 | 31/26/5 | Oral |
| Hammad (2016) | Pediatric systemic lupus erythematosus | Egyptian | 115/259 | PCR-RFLP | 66/44/5 | 141/103/15 | Blood |
| Mucientes (2015) | Panuveitis | Spanish | 340/1815 | Taqman | 139/158/43 | 817/801/197 | Blood |
| Yue (2016) | Myasthenia gravis | Chinese | 475/485 | 48-Plex SNP scanTM Kit | 138/252/85 | 154/249/82 | Blood |
| Marwa (2017) | Rheumatoid arthritis | Tunisia | 108/202 | PCR-RFLP | 74/33/1 | 132/60/10 | Blood |
| Qi (2016) | Graves’ disease | Chinese Han | 700/745 | Taqman | 202/370/128 | 235/357/153 | Blood |
| Shu (2010) | Vogt-Koyanagi-Harada syndrome | Chinese Han | 382/412 | PCR-RFLP | 90/203/89 | 114/214/84 | Blood |
| Shu (2010) | Behcet’s disease | Chinese Han | 362/412 | PCR-RFLP | 111/195/56 | 114/214/84 | Blood |
| Wang (2014) | Multiple sclerosis | Chinese Han | 622/743 | PCR-RFLP | 151/324/147 | 186/386/171 | Blood |
Table 1: Characteristics of studies included in meta-analysis.
| Study | Cases | Controls | Odds ratio | Weight (%) |
|---|---|---|---|---|
| GG/GA/AA | GG/GA/AA | (95% CI) | random | |
| Bogunia-Kubik | 12/44/32 | 20/67/38 | 1.4 (0.6-3.31) | 2.88 |
| Pawlik | 173/193/51 | 118/169/50 | 0.7 (0.44-1.1) | 7.69 |
| Nordang | 246/251/83 | 208/238/58 | 1.21 (0.83-1.77) | 9.51 |
| Nordang | 396/428/114 | 335/461/124 | 0.78 (0.58-1.04) | 12.49 |
| Carvalho | 37243 | 49/20/6 | 0.45 (0.05-4.03) | 0.5 |
| Carvalho | 56/38/6 | 49/20/6 | 0.88 (0.27-2.89) | 1.58 |
| Popovic-Kuzmanovic | 17/26/7 | 18/26/6 | 1.24 (0.35-4.43) | 1.39 |
| Saraiva | 75/39/2 | 31/26/5 | 0.17 (0.03-0.9) | 0.81 |
| Hammad | 66/44/5 | 141/103/15 | 0.71 (0.25-2.04) | 1.99 |
| Mucientes | 139/158/43 | 817/801/197 | 1.28 (0.88-1.87) | 9.72 |
| Yue | 138/252/85 | 154/249/82 | 1.16 (0.79-1.69) | 9.58 |
| Marwa | 74/33/1 | 132/60/10 | 0.18 (0.02-1.42) | 0.55 |
| Qi | 202/370/128 | 235/357/153 | 0.97 (0.72-1.32) | 12.2 |
| Shu | 90/203/89 | 114/214/84 | 1.34 (0.89-2.02) | 8.86 |
| Shu | 111/195/56 | 114/214/84 | 0.69 (0.45-1.05) | 8.33 |
| Wang | 151/324/147 | 186/386/171 | 1.06 (0.78-1.44) | 11.94 |
| Total (fixed effect) | 1964/2610/850 | 2721/3411/1089 | 0.97 (0.87-1.09) | 100 |
| Total (random effects) | 1964/2610/850 | 2721/3411/1089 | 0.97 (0.83-1.14) | 100 |
| Heterogeneity; I2(p) | 32.45% (0.1026) | |||
| Z (p) | Fixed: -0.49 (0.63) | |||
| Random: -0.37 (0.71) | ||||
Table 2: Co-dominant homozygous model (AA vs. GG) of meta-analysis of IL17A rs2275913.
| Study | Cases | Controls | Odds ratio | Weight (%) |
|---|---|---|---|---|
| GG/GA/AA | GG/GA/AA | (95% CI) | random | |
| Bogunia-Kubik | 12/44/32 | 20/67/38 | 1.10 (0.49-2.46) | 1.51 |
| Pawlik | 173/193/51 | 118/169/50 | 0.78 (0.57-1.06) | 7.6 |
| Nordang | 246/251/83 | 208/238/58 | 0.89 (0.69-1.15) | 9.86 |
| Nordang | 396/428/114 | 335/461/124 | 0.79 (0.65-0.96) | 13.21 |
| Carvalho | 37243 | 49/20/6 | 1.63 (0.67-4.0) | 1.25 |
| Carvalho | 56/38/6 | 49/20/6 | 1.66 (0.86-3.23) | 2.19 |
| Popovic-Kuzmanovic | 17/26/7 | 18/26/6 | 1.06 (0.45-2.5) | 1.36 |
| Saraiva | 75/39/2 | 31/26/5 | 0.62 (0.32-1.19) | 2.28 |
| Hammad | 66/44/5 | 141/103/15 | 0.91 (0.58-1.44) | 4.19 |
| Mucientes | 139/158/43 | 817/801/197 | 1.16 (0.91-1.49) | 10.25 |
| Yue | 138/252/85 | 154/249/82 | 1.13 (0.85-1.51) | 8.44 |
| Marwa | 74/33/1 | 132/60/10 | 0.98 (0.59-1.64) | 3.48 |
| Qi | 202/370/128 | 235/357/153 | 1.21 (0.95-1.53) | 10.77 |
| Shu | 90/203/89 | 114/214/84 | 1.20 (0.86-1.68) | 6.82 |
| Shu | 111/195/56 | 114/214/84 | 0.94 (0.68-1.3) | 7.15 |
| Wang | 151/324/147 | 186/386/171 | 1.03 (0.8-1.34) | 9.65 |
| Total (fixed effect) | 1964/2610/850 | 2721/3411/1089 | 0.99 (0.91-1.07) | 100 |
| Total (random effects) | 1964/2610/850 | 2721/3411/1089 | 0.99 (0.90-1.11) | 100 |
| Heterogeneity; I2 (p) | 26.82% (0.1537) | |||
| Z (p) | Fixed: -0.253 (0.80) | |||
| Random: -0.031 (0.98) | ||||
Table 3: Co-dominant heterozygous model (GA vs. GG) of meta-analysis of IL17A rs2275913.
| Study | Cases | Controls | Odds ratio | Weight (%) |
|---|---|---|---|---|
| GG/GA/AA | GG/GA/AA | (95% CI) | Random | |
| Bogunia-Kubik | 12/44/32 | 20/67/38 | 1.21 (0.56-2.62) | 1.68 |
| Pawlik | 173/193/51 | 118/169/50 | 0.76 (0.57-1.02) | 7.71 |
| Nordang | 246/251/83 | 208/238/58 | 0.95 (0.75-1.22) | 9.73 |
| Nordang | 396/428/114 | 335/461/124 | 0.78 (0.65-0.95) | 12.33 |
| Carvalho | 37243 | 49/20/6 | 1.36 (0.58-3.21) | 1.39 |
| Carvalho | 56/38/6 | 49/20/6 | 1.48 (0.80-2.75) | 2.51 |
| Popovic-Kuzmanovic | 17/26/7 | 18/26/6 | 1.09 (0.48-2.48) | 1.5 |
| Saraiva | 75/39/2 | 31/26/5 | 0.55 (0.29-1.02) | 2.45 |
| Hammad | 66/44/5 | 141/103/15 | 0.89 (0.57-1.38) | 4.37 |
| Mucientes | 139/158/43 | 817/801/197 | 1.18 (0.94-1.50) | 10.02 |
| Yue | 138/252/85 | 154/249/82 | 1.14 (0.86-1.50) | 8.44 |
| Marwa | 74/33/1 | 132/60/10 | 0.87 (0.53-1.43) | 3.62 |
| Qi | 202/370/128 | 235/357/153 | 1.14 (0.91-1.42) | 10.47 |
| Shu | 90/203/89 | 114/214/84 | 1.24 (0.90-1.71) | 7 |
| Shu | 111/195/56 | 114/214/84 | 0.87 (0.63-1.18) | 7.28 |
| Wang | 151/324/147 | 186/386/171 | 1.04 (0.81-1.33) | 9.51 |
| Total (fixed effect) | 1964/2610/850 | 2721/3411/1089 | 0.98 (0.91-1.06) | 100 |
| Total (random effects) | 1964/2610/850 | 2721/3411/1089 | 0.99 (0.90-1.10) | 100 |
| Heterogeneity; I2 (p) | 34.63% (0.0853) | |||
| Z (p) | Fixed: -0.491 (0.623) | |||
| Random: -0.242 (0.809) | ||||
Table 4: Dominant model (AA+GA vs. GG) of meta-analysis of IL17A rs2275913.
| Study | Cases | Controls | Odds ratio | Weight (%) |
|---|---|---|---|---|
| GG/GA/AA | GG/GA/AA | (95% CI) | Random | |
| Bogunia-Kubik | 12/44/32 | 20/67/38 | 1.31 (0.73-2.33) | 3.89 |
| Pawlik | 173/193/51 | 118/169/50 | 0.80 (0.53-1.22) | 6.73 |
| Nordang | 246/251/83 | 208/238/58 | 1.28 (0.90-1.84) | 8.61 |
| Nordang | 396/428/114 | 335/461/124 | 0.89 (0.68-1.17) | 12.73 |
| Carvalho | 37243 | 49/20/6 | 0.38 (0.04-3.32) | 0.31 |
| Carvalho | 56/38/6 | 49/20/6 | 0.73 (0.23-2.37) | 1.03 |
| Popovic-Kuzmanovic | 17/26/7 | 18/26/6 | 1.19 (0.37-3.84) | 1.03 |
| Saraiva | 75/39/2 | 31/26/5 | 0.20 (0.04-1.06) | 0.51 |
| Hammad | 66/44/5 | 141/103/15 | 0.74 (0.26-2.09) | 1.3 |
| Mucientes | 139/158/43 | 817/801/197 | 1.19 (0.84-1.69) | 8.85 |
| Yue | 138/252/85 | 154/249/82 | 1.07 (0.77-1.50) | 9.6 |
| Marwa | 74/33/1 | 132/60/10 | 0.18 (0.02-1.42) | 0.34 |
| Qi | 202/370/128 | 235/357/153 | 0.87 (0.67-1.13) | 13.42 |
| Shu | 90/203/89 | 114/214/84 | 1.19 (0.85-1.66) | 9.46 |
| Shu | 111/195/56 | 114/214/84 | 0.72 (0.49-1.04) | 8.14 |
| Wang | 151/324/147 | 186/386/171 | 1.04 (0.81-1.33) | 14.06 |
| Total (fixed effect) | 1964/2610/850 | 2721/3411/1089 | 0.97 (0.88-1.08) | 100 |
| Total (random effects) | 1964/2610/850 | 2721/3411/1089 | 0.98 (0.87-1.10) | 100 |
| Heterogeneity; I2 (p) | 18.64% (0.2404) | |||
| Z (p) | Fixed: -0.546 (0.585) | |||
| Random: -0.393 (0.694) | ||||
Table 5: Recessive model (AA vs. GG+GA) of meta-analysis of IL17A rs2275913.
| Study | Cases | Controls | Odds ratio | Weight (%) |
|---|---|---|---|---|
| G/A | G/A | (95% CI) | random | |
| Bogunia-Kubik | 68/108 | 107//143 | 1.19 (0.80-1.76) | 2.97 |
| Pawlik | 539/295 | 405/269 | 0.82 (0.67-1.02) | 7.55 |
| Nordang | 743/417 | 654/354 | 1.04 (0.87-1.24) | 9.19 |
| Nordang | 1220/656 | 1131/709 | 0.86 (0.75-0.98) | 11.85 |
| Carvalho | 48/14 | 118/32 | 1.08 (0.53-2.19) | 1.02 |
| Carvalho | 150/50 | 118/32 | 1.23 (0.74-2.04) | 1.92 |
| Popovic-Kuzmanovic | 60/40 | 62/38 | 1.09 (0.62-1.92) | 1.55 |
| Saraiva | 189/43 | 88/36 | 0.56 (0.33-0.93) | 1.89 |
| Hammad | 176/54 | 385/133 | 0.89 (0.62-1.28) | 3.4 |
| Mucientes | 436/244 | 2435/1195 | 1.14 (0.96-1.35) | 9.47 |
| Yue | 528/422 | 557/413 | 1.08 (0.90-1.29) | 8.97 |
| Marwa | 181/35 | 324/80 | 0.78 (0.51-1.21) | 2.48 |
| Qi | 774/626 | 827/663 | 1.01 (0.87-1.17) | 10.96 |
| Shu | 383/381 | 442/382 | 1.15 (0.95-1.40) | 8.13 |
| Shu | 417/307 | 442/382 | 0.85 (0.7-1.04) | 7.93 |
| Wang | 626/618 | 758/728 | 1.03 (0.88-1.20) | 10.71 |
| Total (fixed effect) | 6538/4310 | 8853/5589 | 0.98 (0.93-1.04) | 100 |
| Total (random effects) | 6538/4310 | 8853/5589 | 0.98 (0.91-1.06) | 100 |
| Heterogeneity; I2 (p) | 36.96% (0.0687) | |||
| Z (p) | Fixed: -0.625 (0.532) | |||
| Random: -0.471 (0.637) | ||||
Table 6: Allelic model (A vs. G) of meta-analysis of IL17A rs2275913.
In co-dominant homozygous only one OR showed significant association (OR=0.17, 95% CI=0.03-0.9). One study showed significant association (OR=0.79, 95% CI=0.65-0.96) in co-dominant heterozygous model. Similarly in dominant model only one study was found to be with significant OR (OR=0.78, 95% CI=0.65-0.95). None of the ORs reached significance under recessive model. Two studies showed significance in allelic model (OR=0.86, 95% CI=0.75-0.98 and OR=0.56, 95% CI=0.33- 0.93) [25].
The heterogeneity analysis showed non-significant Q values for co-dominant homozygous (Q=22.20, p=0.10), co-dominant heterozygous (Q=20.50, p=0.15) and recessive (Q=18.44, p=0.24) models. Significant Q values were found for dominant (Q=22.95, p=0.09) and allelic models (Q=23.79, p=0.07). The I2 values showed low degree of heterogeneity for co-dominant heterozygous (I2=26.82%) and recessive (I2=18.64%) models; while moderate for co-dominant homozygous (I2=32.45%), dominant (I2=34.63%) and allelic (I2=36.96%) models [26].
Publication Bias
Funnel plots were constructed for each of the co-dominant homozygous, co-dominant heterozygous, dominant, recessive and allelic models. Analysis did not reveal any publication bias and plots were found to be symmetric (Figure 2) [27].
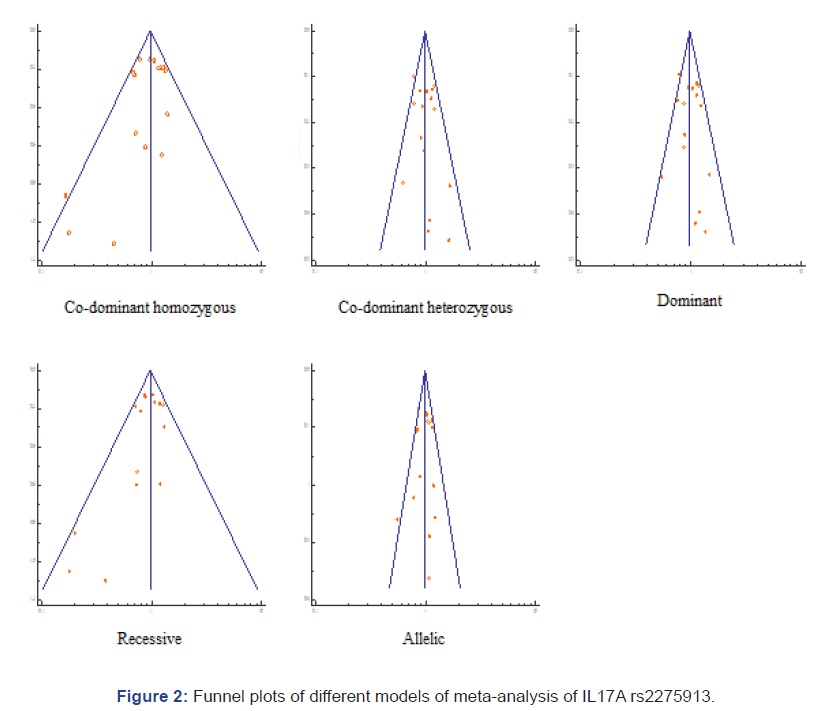
Figure 2: Funnel plots of different models of meta-analysis of IL17A rs2275913.
Interleukin 17A, the founding member of IL17 family of cytokines is one of the key pro-inflammatory mediators exhibiting its role in a variety of ways. Increased levels of IL17A have been found in different autoimmune diseases such as in the synovial fluid of rheumatoid arthritis patients [28].
The rs2275913 SNP is located in the proximal promoter of IL17A gene near two binding sites for the transcription factor nuclear factor of activated T cells (NFATC) which are important in the regulation of IL17A expression [29].
The rs2275913 SNP of IL17A gene has been widely studied for association with autoimmune diseases in different populations due to the pro-inflammatory responses of IL17A cytokine that participate in the pathogenic mechanisms of many autoimmune diseases. However, results have been largely inconsistent with less generalized conclusions [30].
In this meta-analysis we have analyzed 5424 cases and 7221 controls from 16 eligible studies from 13 different research papers for association with AIDs [31].
Meta-analyses of this SNP have been previously performed with respect to cancer risk. In their respective meta-analyses, and found a significant increase in cancer risk associated with rs2275913 SNP. Results of present meta-analysis show no significant AIDs risk modulation with this SNP [32].
Visual inspection of funnel plots revealed no publication bias. The inconsistency (I2) index revealed low degree of heterogeneity (<25%) for co-dominant heterozygous and recessive models. The co-dominant homozygous, dominant and allelic models had moderate degree of heterogeneity (<50%). None of the models showed high degree of heterogeneity (>75%) [33].
There are few limitations of this study. First, the respective number of studies for individual autoimmune diseases was less, therefore a disease-wise subgroup analysis could not be performed. Second, the adjusted analysis by co-variates was not performed due to non-availability of information on confounding factors such as life style and diet [34].
The strengths of the present analysis include a good number of eligible studies and to the best of our knowledge, a first meta-analysis on rs2275913 SNP of IL17A gene with respect to AIDs. Future analyses with greater number of studies will further clarify the role of this SNP in susceptibility to autoimmune diseases in general [35].
To the best of our knowledge, this is the first study to analyze the association of rs2275913 SNP of IL17A gene with autoimmune diseases in a single meta-analysis. The total ORs did not show any significant association for autoimmune diseases. Significant association with reduced susceptibility was seen for periodontitis and rheumatoid arthritis in individual studies.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Benish Ali SH Maigoro AY, (2022) Role of Interleukin 17-A Rs2275913 Polymorphism in Susceptibility to Autoimmune Diseases: A Meta-Analysis. Eur Exp Bio. 12:141.
Copyright: © Maigoro AY, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.