Case Report - (2018) Volume 3, Issue 1
Thiagarajan D*
Diabetes Research Program, Division of Endocrinology, Diabetes and Metabolism, Department of Medicine, New York University Langone Medical Center, New York, USA
*Corresponding Author:
Dr. Devi Thiagarajan Ph.D.,
Diabetes Research Program, Division of Endocrinology, Diabetes and Metabolism
Department of Medicine, New York University Langone Medical Center
New York, USA.
Tel: +1 646 501 7070
E-mail: devithiagarajan@gmail.com
Received date: March 02, 2018; Accepted date: March 09, 2018; Published date: March 12, 2018
Citation: Thiagarajan D (2018) Role of RN Autophagy Mediated ERV Suppression in Cellular Senescence. Insights Biomed. Vol.3 No.1:4
Cellular senescence is defined as a self-imposed, cell cycle proliferation arrest, when cells are subjected to stress encompassing genotoxic, oxidative, replicative damage etc. Senescent cells, secrete plethora of factors including cytokines and chemokines, collectively referred as senescence associated secretory phenotype (SASP). SASP aids in clearing up the damaged cells, by recruiting immune cells, known as senescence surveillance. Several studies have reported enhanced expression of endogenous retrovirus (ERVs) in senescence. Due to their bidirectional transcription, ERVs are capable of eliciting interferon response, assisting in immune clearance. In turn, failure of senescence surveillance leads to accumulation of senescent cells, culminating in functional decline and aging. Though ERVs play an essential role in immune clearance, their role in evading surveillance have not been explored. This study examined the possibility of ERV suppression as an escape mechanism of surveillance. The results indicate that prolonged senescence suppresses ERVs via RNautophagy. Autophagy inhibitor, chloroquine (CQ), activates the ERV expression. In conclusion, this study implicates the role of RNautophagy in ERV suppression as a probable mechanism to escape immune surveillance.
Keywords
Senescence; Endogenous retrovirus; Senescence surveillance; RN autophagy; Chloroquine
Introduction
Cellular senescence is a permanent state of cell cycle arrest in response to stress, including DNA damage, telomeric erosion, oncogenic activation, mitochondrial dysfunction and replicative stress [1,2]. Accumulation of senescent cells contribute to decline of the regenerative potential and function of tissues, accelerates aging. Senescence have been implicated in aging as well as in aging related pathologies including cancer, metabolic syndrome, neurological disorders like Alzheimer’s and Parkinson disease [3]. Hence, senescence is viewed as an essential process to eliminate damaged cells by inducing tissue remodeling [3]. Hallmarks of senescent cells include enhanced senescence associated β- gal (SA-β gal) activity, expression of senescence markers (p16, p21, p19 and pRB) and secretion of a plethora of factors including cytokines, chemokines, matrix metalloproteinases, popularly known as senescence associated secretory phenotype (SASP) [4]. SASP, aid in clearance of senescent cells through both innate and adaptive immune system, termed as senescence surveillance [5].
Transposable elements (TEs) comprise ~50% of human genome and often considered as detrimental, because of their inherent mobile nature [6]. Endogenous retrovirus (ERV) are specialized transposable elements that constitute 8% of human genome and have the ability to transpose through DNA or RNA intermediates [7,8]. Though majority of ERVs have lost their ability to actively move across the genome, some may still impact their host by modulating nearby genes. Several human diseases have been linked to ERVs, including cancer, amyotropic lateral sclerosis (ALS), multiple sclerosis (MS) and schizophrenia [9].
Several studies have successfully demonstrated increased ERV expression during senescence and aging [10-12]. Bidirectional transcription of many TEs including ERVs results in dsRNA [13,14]. dsRNAs, thus formed trigger interferon response, facilitating immune clearance of ERV-activated cells [15,16]. Thus, increased ERV could potentiate senescent cell clearance.
Recent work from our laboratory showed ERV dysregulation in acute myeloid leukemia (AML) development. ERVs were increased during early myeloid dysplastic syndrome (MDS) and were significantly suppressed in high risk MDS. Also, it suggests ERV suppression as a potential mechanism in modulating immunogenicity regarding AML development [6].
Current study aimed in identifying the role of ERVs in prolonged senescence. Interestingly, results indicate that during prolonged senescence, ERVs are suppressed via specialized autophagy, RNautophagy. Hence, suggesting ERV suppression as a prospective mechanism to evade immune surveillance.
Case Study
All animal procedures were approved by the Institutional Animal Care and Use Committee at New York University School of Medicine. All mice studied were male, had free access to water and were subjected to 12 h light/dark cycles. WT C57BL/6 male that were 2 months old, were used for the study. For primary skin fibroblasts, 2 mm ear lobe tissues were minced in DMEM buffer containing Type III Collagenase overnight and passed through 50μm filter to obtain single cell suspension. The attached cells were then cultured and passaged in DMEM supplemented with 10% fetal bovine serum (FBS). HCT116 and IMR-90 cells were purchased from ATCC and maintained in McCoy5A and EMEM medium, supplemented with 10% FBS. Media and FBS were purchased from GIBCO.
Mitomycin C, was purchased from SigmaAldrich. Cells were treated with and without 10 μM MMC for 4hrs, washed and incubated in fresh media. Cells were analyzed 4 or 12 days post treatment.
RNA was isolated using Qiagen RNAesy Kit (Qiagen). cDNA was synthesized using superscript cDNA synthesis kit (Invitrogen). qPCR performed using FastSybr green mastermix (Applied biosystems) and were normalized against β-actin. Table 1 lists the primer sequences used.
| Mice Primers Name | Sequences |
|---|---|
| MICE p21 FP | ACGGGACCGAAGAGACAAC |
| MICE p21 RP | CAGATCCACAGCGATATCCA |
| MICE p16 FP | TCGAATCTGCACCGTAGTTG |
| MICE p16 RP | CGTGAACATGTTGTTGAGGC |
| MICE LINC9 FP | TGAGTGGAACACAACTTCTGC |
| MICE LINC9 RP | CAGGCAAGCTCTCTTCTTGC |
| MICE MULV FP | TTCTGCTCCTCTTCTGCCCT |
| MICE MULV RP | GAGGACCCTGGGCAAGAAAC |
| MICE MMTV FP | TTTCCCGAAGAAGGAGGATT |
| MICE MMTV RP | GCTTCTGCGGATAGCAAAAC |
| MICE MUSD FP | ATAGAGGCCGCTTCTTTGC |
| MICE MUSD RP | TGAGACTCCACCAAATGTCC |
| MICE ACTIN FP | AGGCCAACCGTGAAAAGATG |
| MICE ACTIN RP | GCTGAGAAGCTGGCCAAAGA |
| Human Primers Name | Sequences |
| human p21 RT FP | CATGGGTTCTGACGGACAT |
| human p21 RT RP | AGTCAGTTCCTTGTGGAGCC |
| human p16 RT FP | GGGTCGGGTGAGAGTGG |
| human p16 RT RP | CGAATAGTTACGGTCGGAGG |
| human ERV K RT FP | CACAACTAAAGAAGGCTGACG |
| human ERV K RT RP | CATAGGCCCAGTTGGTATAG |
| human ERV L RT FP | CTTCAGCTGGCAAGGCC |
| human ERV L RT RP | CCAGTGTGATATCTTGTGGC |
| human ERV W RT FP | TGAGTCAATTCTCATACCTG |
| human ERV W RT RP | AGTTAAGAGTTCTTGGGTTGG |
| human ERV R RT FP | CATGGGAAGCAAGGGAACT |
| human ERV R RT RP | CTTTCCCCAGCGAGCAATAC |
| human Actin RT FP | CATGTACGTTGCTATCCAGGC |
| human actin RT RP | CTCCTTAATGTCACGCACGAT |
Table 1: Sybr green primers (Source: Life technologies).
For lysates, cells were suspended in RIPA buffer (25 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% TritonX100), with protease inhibitor cocktail. 40 μg lysate were loaded on 4-15% Bis-Tris gel and blotted onto nitrocellulose membrane. Blots were probed for BNIP3, BECLIN1, ATG5, ATG7, LCIII, p-mTOR and t-mTOR (Cell Signaling Technologies), β-ACTIN (Santacruz), LAMP2B and SIDT2 (Abcam). Bands were normalized to β-ACTIN and quantified using ImageJ.
Data were analyzed using Graphpad Prism software. All data are expressed as mean± standard error mean (SEM). Results were analyzed using two-tailed unpaired t-test. P values < 0.05 were considered significant.
Results and Discussion
Primary dermal fibroblasts, isolated from 2 months old WT C57BL/6 mice were used in this study. Primary mice fibroblasts were treated with either vehicle or mitomycin C (MMC), to induce senescence [17]. Cells were analyzed 4 and 12 days post treatment. qPCR for senescence markers (p21 and p16), revealed increased expression of both the markers studied (Figure 1A). Results indicate the induction of senescence upon MMC treatment. Further, ERVs (Merv-k, MusD, MuLV and Linc9) were examined via qPCR in mock vs senescent fibroblasts. As can be seen in (Figure 1B), ERVs were upregulated in 4 days post senescence (Sen 4 d), which then were then downregulated in 12 days post senescence (sen 12 d).

Figure 1: ERVs are upregulated in senescence.
To verify the results, the same experiment was repeated in two different human cell lines- IMR-90 (human diploid fibroblasts) and HCT116 (colorectal cancer cell line). Both the cell lines were induced senescence using MMC and senescence as well as ERV expression were analyzed via qPCR. The results consistently showed ERV downregulation at 12 days post senescence, senescent post 12 days, compared to 4 days post senescence, despite no decrease in the senescence markers, p16, p21 (Figures 2A-2D).
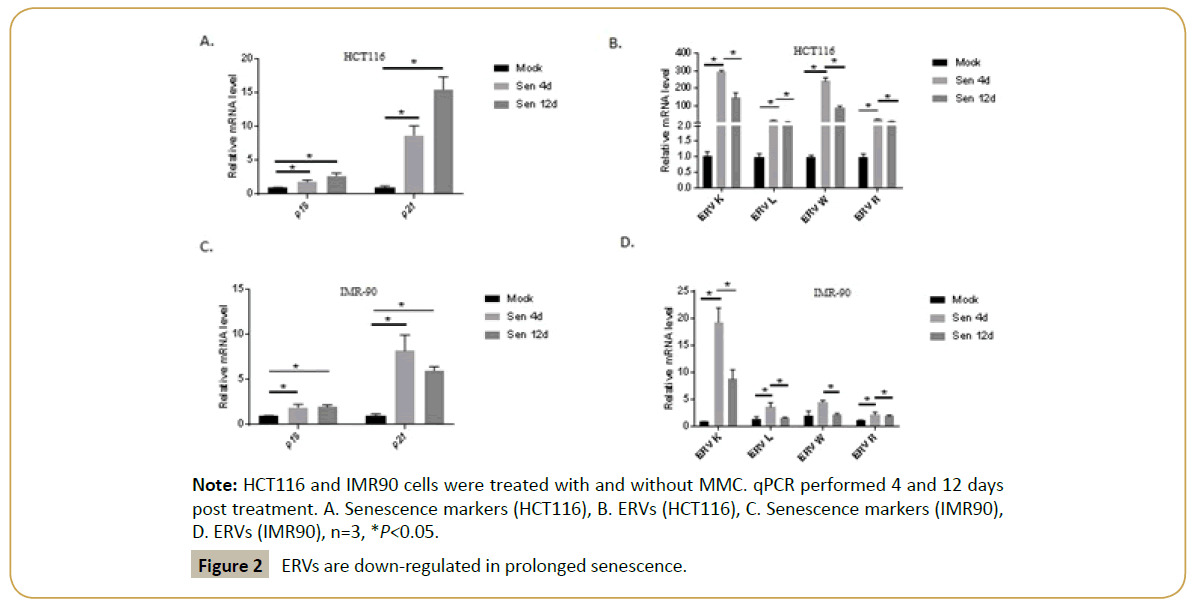
Figure 2: ERVs are down-regulated in prolonged senescence.
Lysosomes degrade various biomolecules including nucleic acids through autophagy. Autophagy mainly consist of three types namely, micro, macro and chaperone mediated. Recent studies suggested the fourth autophagic mechanism, RNautophagy, to degrade retrotransposon RNA [18-20]. To examine whether RNautophagy was activated in prolonged senescence condition, lysates were made from primary mice fibroblasts, in mock, 4 and 12 days post senescence. Western blot for autophagy related proteins including LCIII, ATG5, ATG7, mTOR, BNIP3, BECLIN1, LAMP2B and SIDT2 were carried out. Results indicate that key autophagic proteins including LCIII, mTOR, and RNautophagy related proteins-LAMP2B, SIDT2 were suppressed at 4 days post senescence, while were significantly upregulated in 12 days post senescence (Figure 3A).
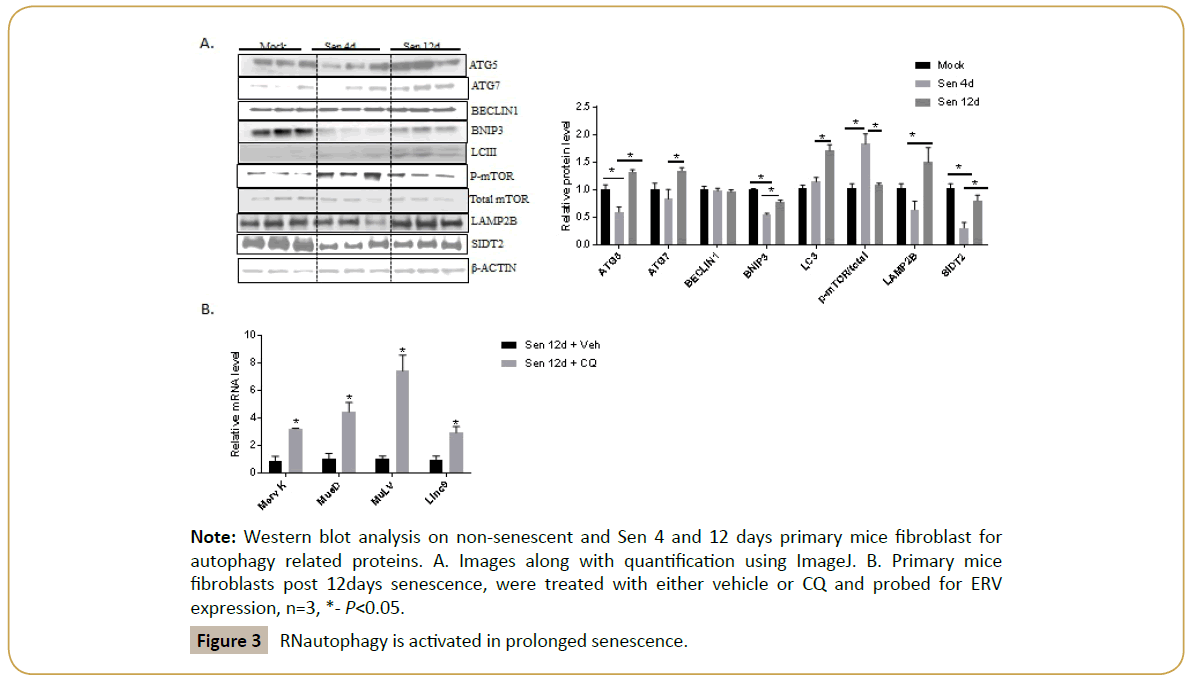
Figure 3: RNautophagy is activated in prolonged senescence.
To further examine, whether autophagy is responsible for ERV suppression, cells were treated with chloroquine (CQ), an autophagy inhibitor 12 d post treatment. When compared to vehicle treated post senescent 12-day fibroblasts, CQ treatment drastically enhances the ERV expression (Figure 3B). This data clearly demonstrates the role of autophagy in ERV suppression.
Senescence is viewed as an essential process for tissue remodeling. While acute senescence aids in wound healing and tissue repair, under chronic settings, senescence contributes to functional decline and aging [3]. Senescent cells, through ERVs and SASP, recruit immune cells, to eliminate them. However, prolonged senescence is the result of evasion of surveillance. This study demonstrates ERV suppression via RNautophagy, as an escape mechanism for immune surveillance.
Increased ERV expression can lead to genomic instability, resulting in cancer development and cancer occurs when cells bypass senescence [8]. However, our results suggest ERV suppression aids in evading surveillance, resulting in senescent cells accumulation, which seems to be detrimental. Nonetheless, just like senescence, ERV expression under acute settings, assists in clearing damaged cells, while chronic expression leads to genomic instability and cancer. Like most of the biological phenomenon, senescence and ERV regulation seems to be a ‘double edged’ sword.
Autophagy is a catabolic process, where macromolecules are degraded by lysosomes. Autophagy occurs at basal level in normal condition but is accelerated during stress and is essential for biological homeostasis. Apart from the macroautophagy, microautophagy and chaperone mediated autophagy, RNA/ DNAutophagy forms the fourth type, where DNA and RNA are degraded by lysosomes in ATP dependent manner [18,20]. Loss of autophagy has been shown to cause premature aging in many species [21,22]. Treatment with transcription factor EB (TFEB), a master regulator of lysosomal biogenesis has shown to extend life span in worms and lessen the metabolic syndrome in mice [23].
Conclusion
Several evidences are in favor of autophagy activation to extend longevity. Autophagy activation have been shown to induce senescence [24]. Our results show, that autophagy suppression might help the senescent cells to escape surveillance and contribute to aging. Nevertheless, it is the type of autophagy cargo, which determines the outcome in terms of longevity and aging. This study is in lieu with the observations in AML patients, where ERV regulation correlates with Lamp2, RNautophagy marker [6]. Senolytic drugs under development targets these surveillance resistant senescent cells. Hence, mechanism mediating evasion from senescence surveillance is essential for designing better senolytic drugs and this study provides a possible direction.