Joseph Omololu-Aso*, Oluwaseun Oluwatoyin Omololu-Aso, Micheal T Atiene, Adekunle Adejuwon, Alexandrer Tuesday Owolabi and Arwa Shesha
Department of Microbiology, Obafemi Awolowo University Ile-Ife, Osun Nigeria
Corresponding Author:
Joseph Omololu-Aso
Obafemi Awolowo University Ile-Ife, Osun Nigeria
Tel: +234-8033770933
E-mail: omololuaso@oauife.edu.ng
Received date: February 04, 2017; Accepted date: February 10, 2017; Published date: February 15, 2017
Citation: Omololu-Aso J, Omololu-Aso OO, Atiene MT, et al. Salmonellosis and Shigellosis Associated with Cattle Dung Contaminant from Indigenous Abattoirs, Osun State, Nigeria. Br J Res 2017, 4:2. doi: 10.21767/2394-3718.100002
Copyright: © 2017 Omololu-Aso J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
Salmonella-Shigella ; Fecal samples; Antibiotics; Abattoir; Infection; Contaminant
Introduction
Salmonella are the leading cause of bacteremia, 28.5% with S. typhi accounting for 20.9% and non-typhi Salmonella 7.6% [1]. Infections due to Salmonella serotypes continue to be a major health problem [2]. Worldwide incidence of shigellosis is estimated to be 164.7 million cases per year of which 163.2 million were in developing countries, where 1.1 million deaths occurred. About 60% of all episodes and 61% of all deaths attributable to shigellosis involved cattle and children below five years. The incidence in developing countries may be 20 times greater than that in developed countries [3]. An estimated 30% incidence of these infections are caused by S. dysenteriae with case fatality rates reaching 30% [3].
Salmonellosis is one of the most common and widely distributed food-borne diseases. It constitutes a major public health burden and represents a significant cost in many countries [4]. Most Salmonella infections in cattle are subclinical, and shedding of the organism can occur for extended periods. Salmonella can also persist in the farm environment [5].
Shigella species are highly infective, particularly S. dysenteriae considered the most virulent, and can produce a potent cytotoxin known as “Shiga toxin”. Shigella dysenteriae type 1 causes severe and sometimes fatal disease. Shigella species have been recently identified to be the most frequently identified agent of laboratory-acquired infections because of their high virulence and low infectious dose [6].
In Nigeria, morbidity associated with illnesses due to Salmonella continues to be on the increase and in some cases resulting in deaths [7]. It is common in developing countries where affected populations are immunologically compromised due to poor nutrition and background infections leading to high morbidity attributed to shigellosis [8-10]. Salmonellosis, a major cause of bacteria enteric illness in both humans and animals cause diseases in humans and is referred to as a zoonotic infection/disease. It is a disease caused by the bacteria Samonella [4].
The aim of this research work was to isolate, identify and characterize Salmonella and Shigella from fecal contaminants on products and the antimicrobial susceptibility of the isolates obtained from the abattoirs.
Materials and Methods
Study area
The study area was Ife Central Local Government, Ile-Ife, Nigeria. Samples tested include the fresh cattle dung contaminant from separate meat products at various butcheries which were collected between February to June, 2016. Ife Central is a local government area in Osun State, Nigeria. Its headquarters are in the city of Ile-Ife to the south of the area. It has an area of 111km2 and a population of 167,254 at 2006 census.
Sample collection and preparation
Each sample consisted of cattle dung contaminant collected with sterile applicator swabs. The various samples collected were from Odo-Eran area along Ede Road, as well as from an abattoir adjacent to God’s Love Terbanacle, along Ibadan Road. All of the samples collected were from the dung of freshly killed cows.
Sample transportation
The samples were placed within sterile plastic containers and transported to the Microbiology Laboratory, Obafemi Awolowo University, Ile-Ife, Nigeria within an hour of collection. The samples were examined immediately upon arrival at the laboratory. Each Sample was placed aseptically in sterile nutrient broth and incubated.
Microbiological analysis of samples
The samples collected were serially diluted and analyzed on Salmonella-Shigella agar by the direct plating method. A measured amount of the moderately warm molten agar was poured into the sterile petri dishes and allowed to set after which the organisms were streaked on the Salmonella-Shigella agar plates evenly to allow for the spread of the organism. The plates were then incubated for 24-48 hours at 37°C in an inverted manner. Biochemical tests were employed to confirm the isolates including Gram staining, catalase, oxidase, urease, glucose fermentation, motility tests. For all the biochemical tests carried out 18-24 hours young cultures of the isolates were used.
Antibiotic susceptibility test
In vitro susceptibility of the isolates against antimicrobial agents was determined by the standard disc diffusion procedure (Bauer et al 1996). The following antibiotic discs were used: augmentin (30 μg), gentamycin (30 μg), streptomycin (30 μg), pefloxacin (10 μg), chloramphenicol (23.2 μg), amoxacillin (25 μg), ciprofloxacin (5 μg), streptomycin (30 μg), tarivid (5μg) spafloxacin (25 μg) and septrin (10 μg). Isolates were inoculated in nutrient broth and incubated at 37°C for 24hours. The inoculum was poured into the Mueller-Hinton agar plates, antibiotic discs were placed on the plates. Plates were incubated at 37°C for 18 to 24 hours. The zones of inhibition were measured, recorded and interpreted according to the Clinical Laboratory Standard Institute provided (CLSI 2014).
Results
Biochemical characterization of isolates of Shigella and Salmonella strains
The isolates were Gram negative rods. They were catalase positive; motility positive for Salmonella but motility negative for Shigella. They were urease positive and glucose positive.
However, Salmonella tested positive in the oxidase test. Shigella tested negative for oxidase (Tables 1 and 2) (Figures 1-4).
| Morphology |
Results |
| Growth in Nutrient agar |
Positive for all isolates |
| Growth in MacConkey agar |
Positive for both Salmonella and Shigella species |
| Growth in Salmonella-Shigella agar |
Positive for Salmonella and Shigella species |
| Motility testing |
Positive for Salmonella species but negative for Shigella species |
| Gram staining |
Negative for both Salmonella and Shigella species |
Table 1: Morphological reaction of isolate to the selected media.
| Antibiotics |
Total Isolates |
R (%) |
I (%) |
S (%) |
| Septrin |
30 |
75 |
25 |
25 |
| Chloramphenicol |
30 |
100 |
0 |
0 |
| Sparfloxacin |
30 |
15 |
30 |
30 |
| Ciprofloxacin |
30 |
25 |
10 |
65 |
| Amoxicillin |
30 |
40 |
30 |
30 |
| Augmentin |
30 |
20 |
0 |
80 |
| Gentamicin |
30 |
35 |
10 |
55 |
| Pefloxacin |
30 |
10 |
60 |
30 |
| Tarivid |
30 |
10 |
60 |
30 |
| Streptomycin |
30 |
35 |
0 |
65 |
Key: Susceptible, I – Intermediate, R – Resistance
Table 2: Antibiotic susceptibility pattern of Salmonella/Shigella species isolated from cattle dung collected from abattoirs in Ile- Ife town, Osun State, Nigeria.
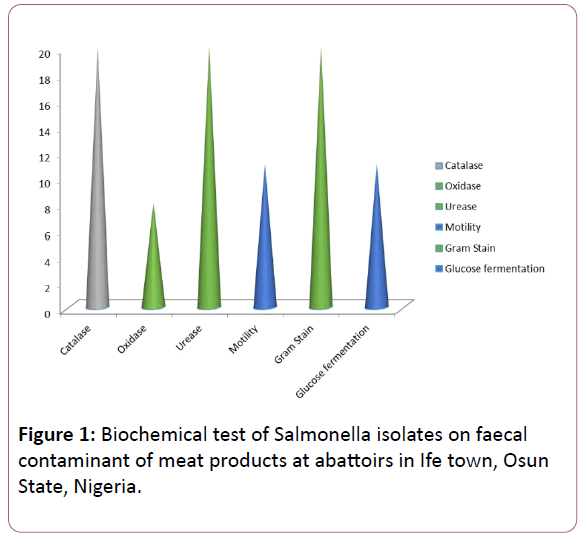
Figure 1: Biochemical test of Salmonella isolates on faecal contaminant of meat products at abattoirs in Ife town, Osun State, Nigeria.
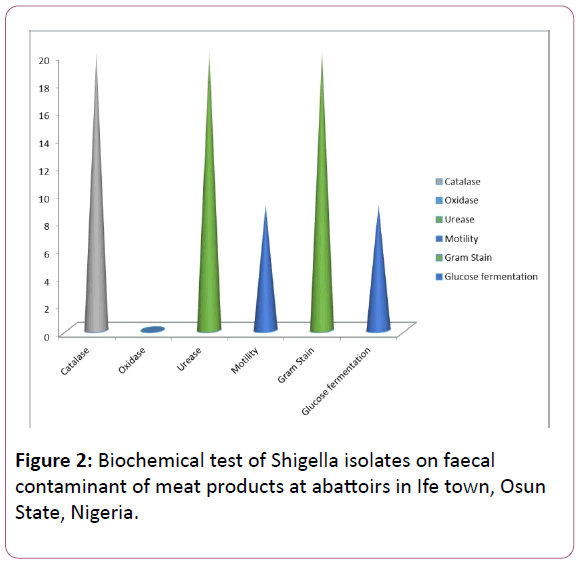
Figure 2: Biochemical test of Shigella isolates on faecal contaminant of meat products at abattoirs in Ife town, Osun State, Nigeria.
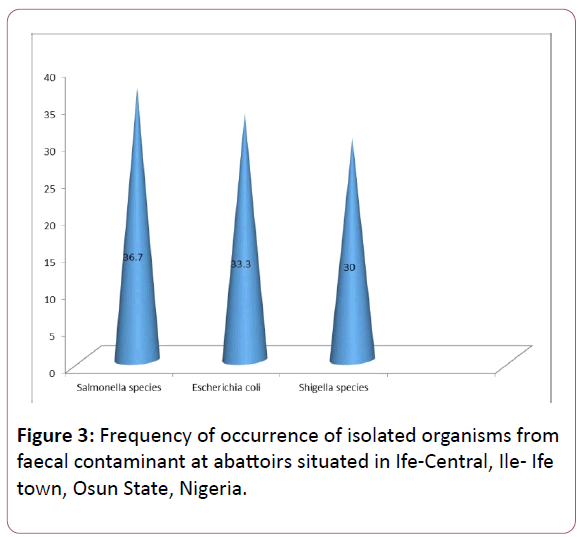
Figure 3: Frequency of occurrence of isolated organisms from faecal contaminant at abattoirs situated in Ife-Central, Ile- Ife town, Osun State, Nigeria.
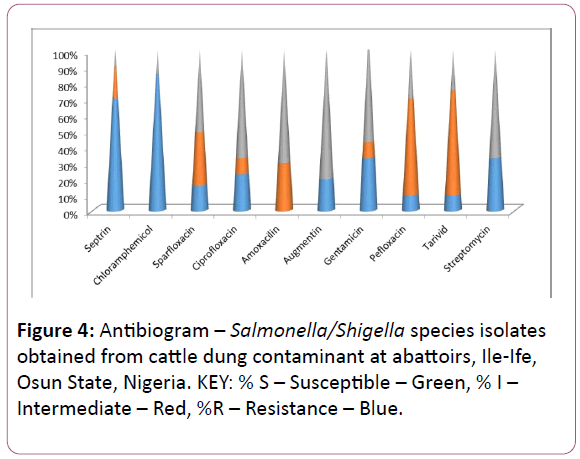
Figure 4: Antibiogram – Salmonella/Shigella species isolates obtained from cattle dung contaminant at abattoirs, Ile-Ife, Osun State, Nigeria. KEY: % S – Susceptible – Green, % I – Intermediate – Red, %R – Resistance – Blue.
Discussion
In this study, out of the ten (10) samples collected in Odo-Eran area in Ife town, 4 (40%) samples showed Escherichia coli growth as seen in Figure 3. 4 (40%) Salmonella were isolated and 2 (20%) Shigella-like organisms were isolated. Thus, the probability of the occurrence of bacterial infection among those that consume meat or work in the butcheries in this community is high and which is in agreement with the reports of Joseph et al. (2012) on Salmonellosis and associated organisms as dangerous food borne pathogen.
From the 20 samples collected in God’s Love Tabernacle (GLT) area in Ife town, 7 (35%) showed Salmonella growth as seen in Figure 1. 7 (35%) confirmed Shigella-like characteristics (Figure 2). Six (30%) isolates left were observed to show Escherichia coli growth as in Figure 3.
All of the results suggest that livestock and in this case cow dung is a reservoir for bacteria for environmental contamination which is in concordance with the findings of Sayah [11].
Table 1 showing the reactions of isolates of Salmonella and Shigella to the various biochemical tests carried out, each indicating the distinctive properties of the isolates such as motility, with Salmonella being motile and Shigella being nonmotile. These tests are effective in determining a basic level and type of bacteria present in the cow dung and hence relating to the type of infection suffered by the cattle.
From our findings, it can be established that, although the level of Salmonella may not be high in these understudy area, there is still a possibility for infection in cases of poor hygienic practices. Also, it can be inferred that the chances of infection remain high among the butchers and meat handlers which corroborated the reports of Hawkin [12] in the clinical management of public health risks from Salmonella.
From the Tables 2 showing the antibiotic susceptibility testing, the Salmonella/Shigella isolates were 100% resistance to chloramphenicol, 75% to septrin and 10% to pefloxacin and tarivid respectively while the isolates were 80% susceptible to gentamicin and 65% to ciprofloxacin and streptomycin used as illustrated in Figure 4. In our findings, it can be inferred that both gentamicin streptomycin and ciprofloxacin antibiotic therapeutic agents will be effective in the treatment of cattle infections caused by Salmonella and Shigella species and which is in variance with the work of Nyogi [13] on changing pattern of serotypes on antimicrobial susceptibility of Shigella sp. Isolated from children in Calcutta. Multiple antibiotic resistances shown by the isolates may suggest that the cattle are reared in areas where antibiotics are often abused [14].
References
- Obaro S (2011) Invasive Salmonellosis in central Nigeria.
- Salehi TZ, Mahzounieh M, Saeedzadeh A (2005) Detection of InVA gene in isolated Salmonella from Broilers by PCR method. International journal of Poultry science 4: 557-559.
- Jaya S, Saurabh G, Sonam M, Bharti A (2014) Evaluation of antimicrobial Activity of Ganoderma lucidium. Int Journal of Advanced Research 2: 460-466.
- WHO (2013) Salmonella (non-typhoidal) fact sheet N°139.
- Huston CL, Wittum TE, Love BC, Keen JE (2002) Prevalence of fecal shedding of Salmonella spp. in dairy herds. Journal of Animal Vet Med Association 220: 645-649.
- Peng J, Yang J, Jin Q (2011) An Integrated approach for finding overlooked genes in Shigella. PLoS One 6: e18509.
- Akinyemi KO, Coker AO, Olukoya DK, Oyefolu AO, Amoroghoye EP, et al (2000) Prevalence of multi-drug resistant Salmonella typhi among clinically diagnosed typhoid fever patients in Lagos, Nigeria. Z Naturforsch 55: 489-493.
- Mammina C, Ranjbar R (2008) “Shigellosis”: disease and epidemiology.
- Delpech V, O’Sullivan B, Pontivivo G, Karagiannis T, Marriott D, et al (2002) Shigellosis linked to sex venues, Australia.
- Woodward DL, Clark CG, Caldeira RA, Ahmed R, Soule G, et al (2005) Identification and characterization of Shigella boydii 20 serovar Nov., a new and emerging Shigella serotype. J Med Microbiol 54: 741-748.
- Sayah RS, John BK, Yvette J, RoseAnn M (2004) Patterns of Antimicrobial Resistance Observed in Escherichia coli Isolates Obtained from Domestic- and Wild-Animal Fecal Samples, Human
- Niyogi SK, Mitra U, Dutta P (2010) Changing pattern of serotypes, antimicrobial susceptibilities of shigella species isolated from children in Calcutta. Journal of Infectious Diseases 54: 121-122.
- Joseph AO, León-Velarde CG (2012) Salmonella Detection Methods for Food and Food Ingredients, Salmonella - A Dangerous Foodborne Pathogen.
- Hawkins LL (2000) Salmonellosis in feed yards: epidemiology, clinical management, and public health risk, Proc. Annu Conv Am Assoc Bovine Pract 33: 133-136.