Blendi Ura1*, Luigina De Leo1, Giorgio Arrigoni2,3, Federica Scrimin1, Irene Berti and Tarcisio Not1,4
1Institute for Maternal and Child Health-IRCCS, Burlo Garofolo, Trieste, Italy
2Department of Biomedical Sciences, University of Padova, Padova, Italy
3Proteomics Center, University of Padova and Azienda Ospedaliera di Padova, Padova, Italy
4University of Trieste, Trieste, Italy
*Corresponding Author:
Blendi Ura
Institute for Maternal and Child Health-IRCCS, Burlo Garofolo, Trieste, Italy
Tel: +39 040 3785863
Fax: +39 040 3785210
E-mail: blendi.ura2006@libero.it
Received date: December 07, 2016; Accepted date: December 27, 2016; Published date: December 30, 2016
Citation: Ura B, Leo LD, Arrigoni G, et al. Serological Proteome Analysis for Identification of Potential Antigen in Atopic Dermatitis. Ped Health Res 2016, 1:1. doi: 10.2176/2574-2817.100004
Objective: Identify human skin derived auto antigens that could be recognized by IgE serum of patients with both severe atopic dermatitis and high serum IgE immunoglobulins.
Design: The study was performed retrospectively on sera collected from children with severe atopic dermatitis with total IgE serum concentration (IgE>3000 U.I./ml), as a serum sample controls we used the children’s sera suffering from severe food allergy and with high IgE concentrations against food proteins (IgE>90 kU/l). All these serum samples were used to identify skin-derived antigen by using immunoproteome technique.
Setting: Institute for Maternal and Child Health.
Patients: Six children suffering from severe atopic dermatitis and six suffering from severe food allergy without skin involvement.
Interventions: Serological proteome analysis (SERPA) for antigen identification.
Main outcome measures: First immunoproteomic study for identification of potential antigen in severe atopic dermatitis. Identification of two immunoreactive proteins IGKC (Ig kappa chain C region) and IGLC1 (Ig lambda-1 chain C regions).
Results: After membranes analysis we identified two immunoreactive proteins IGKC and IGLC1 present only in membrane treated with severe atopic dermatitis. Reaction of these proteins were considered non-specific event and didn’t further investigate.
Conclusions: Using SERPA analysis we identified two immuno-reactive proteins. Even these proteins have proved to be non-specific event; this showed the great potential of SERPA for antigen identification.
Keywords
Atopic dermatitis; Antigen; Serological proteome analysis
Introduction
Atopic dermatitis is a skin disease characterize by an inflammatory component with an possible immunologic disturbance that causes IgE-mediated sensitization [1]. Some studies indicate that patients with atopic dermatitis were primarily sensitized to exogenous allergens such as grass pollen derived calcium-binding proteins [2] or environmental fungal manganese superoxide dismutase [3] and subsequently developed IgE cross-reactivity to self-proteins, perhaps via molecular mimicry mechanisms. Specific IgE antibodies against the human calcium-binding protein (Homs4) and the human manganese superoxide dismutase were found in 10% and 43% of patients respectively with severe atopic dermatitis. Until now no skin derived auto-antigen were identified in atopic dermatitis. Here we present an immunoproteomic study to identify human skin derived auto antigens that could be recognized by IgE serum of patients with both severe atopic dermatitis and high serum IgE immunoglobulins.
Material and Methods
We analyzed the serum samples of 6 children suffering from severe atopic dermatitis (6 M, mean age 10 years) with high total IgE serum concentration (mean value 9373 ± 6232 U.I./ ml), the serum samples were collected before the immunosuppressive therapy. As a control, we analyzed the serum samples of 6 children (6 M, mean age 10 years) suffering from severe food allergy without skin involvement as previously described [4] with high IgE concentrations against food proteins (IgE values>90 kU/l) and a positive history of at least one severe allergic reaction.
2-DE, Western blot and image analysis
About 300 mg of skin, that was obtained, after informed consent from parents, from children during surgery as a result of traumatic injuries involving the skin, was homogenate in 2- DE dissolution buffer. The solutions centrifuged at 10,000 × g at 4°C for 30 min. The protein content of the supernatant was determined using the Bradford assay. Three hundred micrograms of proteins were used for the 2-DE analysis.
NL IPG Ready strips, 7 cm, pH 3-10 were used for IEF. After equilibration IPG strips were transferred onto a 12% polyacrylamide gel for the second dimension. For immunoproteomic the experiments were performed in double replicates while one gel was used for control.
After the second dimension the proteins were transferred to a nitrocellulose membrane in a blotting chamber. The residual binding sites on the membrane were blocked by treatment with 3% TBS-Tween 20, and incubated overnight at 4°C with 1:20 diluted serum mix of patients and in parallel with serum mix of control. After washing the membranes were incubated overnight with 1: 3000 diluted anti-IgE at 4°C. Finally, the membrane was incubated with 1:8000 diluted HRP-conjugated anti-rabbit IgG. At the end the protein presence was visualized by chemiluminescence. The intensity of the signals was quantified by Versa Doc Imaging system while 2-DE gel was scanned with a Molecular Imager Pharos FX system (Bio-Rad) and the analysis of the spots was carried out using the Proteome Weaver 4 program (Bio-Rad).
In gel digestion and MALDI -TOF/TOF analysis
Spots from 2-DE were digested with sequencing grademodified trypsin (Promega, Madison, WI, USA) and analyzed by mass spectrometry, as described by Ura et al. [5]. Briefly, the protein spot was manually excised from the gels with sterile tips, washed several times with 50 mM NH4HCO3 and acetonitrile (ACN). Ten μl of trypsin (12.5 ng/μl in 50 mM NH4HCO3) were added to each sample which kept in ice for 30 min before being incubated overnight at 37°C. For peptide extraction three changes were made by using 75% ACN/0.1% trifluoroacetic acid (TFA) dried under vacuum and then dissolved again in 10 μl of 0.1% TFA. For MALDI analysis 1 μl of the sample was mixed with 1 μl of (matrix α-cyano-4- hydroxycinnamic acid, 5 mg/ml in 70% ACN/ 0.1% TFA) and an amount of 0.8 μl of the final sample/matrix mixture were spotted onto a MALDI target plate. The analysis was performed on a MALDI-TOF/TOF 4800 mass spectrometer (AB Sciex, Framingham, MA, USA) in a data dependent mode: a full MS scan was acquired, followed by MS/MS spectra of the 10 most intense signals.
The MS/MS spectra were converted into Mascot Generic Format (MGF) files and analyzed with Proteome Discoverer software (Thermo Fisher Scientific). The human section of the Uniprot database was used for data searching: tolerance was set to 50 ppm and to 0.3; enzyme specificity was set to trypsin with up to one missed cleavage. Carbamidomethylation of cysteine residues was set to fixed modification and methionine oxidation to variable modification.
The software calculates the FDR (False Discovery Rate) based on the search against a randomized database: proteins were considered as positively identified if at least two unique peptides were identified for each protein with confidence (q<0.05).
Results
For SERPA (Serological Proteome Analysis) membrane analysis we used two criteria to eliminate the false positive: 1) The immunoreactive spots should be present only in membrane treated with eczema sera. 2) The immunoreactive spots should be present in all experiments.
After fixing the criteria the membranes treated with severe food allergy without skin involvement sera were marched with the membranes treated with eczema sera for difference identification using the Proteome Weaver 4. Once find the differences we matched the membrane treated with eczema sera with the 2-DE gel for protein spot in gel position.
The analysis revealed 2 immunoreactive spots in membrane treated with eczema sera. The protein spots were identified by MALDI-TOF/TOF (Table 1 and Figure 1) and correspond to IGKC (Ig kappa chain C region) and IGLC1 (Ig Lambda-1 chain C regions). Reaction of these proteins were considered nonspecific event and didn’t further investigate.
| Accession Number |
Spot no. |
Protein Description |
Gene Symbol |
Protein Score |
Molecular weight |
Peptides |
| A0A087X1V9 |
1 |
Ig kappa chain C region |
IGKC |
174 |
25.950 |
K.SGTASVVCLLNNFYPR.E
K.VYACEVTHQGLSSPVTK.S
R.TVAAPSVFIFPPSDEQLK.S |
| P0CG04 |
2 |
Ig lambda-1 chain C regions |
IGLC1 |
85 |
11.512 |
K.YAASSYLSLTPEQWK.S
K.ATLVCLISDFYPGAVTVAWK.A |
Table 1: List of immunoreactivity proteins as identified by the MALDI-TOF/TOF mass spectrometry.
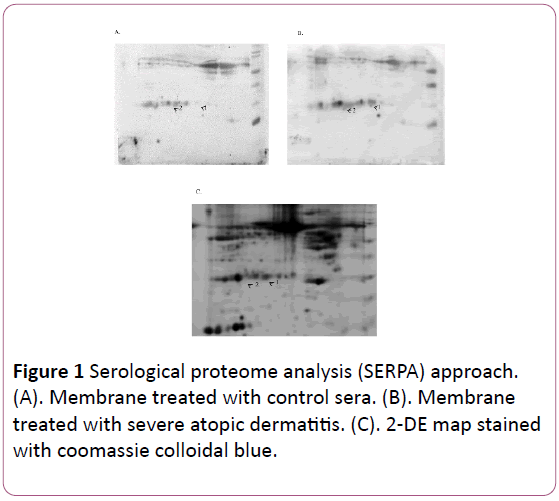
Figure 1: Serological proteome analysis (SERPA) approach. (A). Membrane treated with control sera. (B). Membrane treated with severe atopic dermatitis. (C). 2-DE map stained with coomassie colloidal blue.
Discussion and Conclusion
We conducted this work based on the review [1] in which is assumed the presence of antigens against IgE in atopic dermatitis. Best of our knowledge this is the first immunoproteomic study conduct on atopic dermatitis for identification of antigen against IgE using whole skin proteome.
SERPA has been used for identification of immunoreactive proteins in various diseases [6]. Unfortunately, we could not identified a valid antigen by SERPA analysis. This may be correlated with analytical limitations inherent in 2-DE: First, 2- DE is limited to identifying relatively abundant proteins limiting the sensibility. Second, 2-DE is limited in the separation of certain proteins like plasma membrane proteins, which are of great interest for the antigens search [7]. Although with several limit SERPA remain the method of choice for identification of antigens.
For future prospective SERPA analysis should be addressed on disease skin as antigen resource. Another important aspect is the sub-proteome (especially plasma membrane proteins) to reduce the proteome complexity and to permit deep investigation for antigen identification.
References
- Bieber TH (2008) Atopic dermatitis. N Engl J Med 358: 1483-1494.
- Valenta R, Duchêne M, Pettenburger K, Sillaber C, Valent P, et al. (1991) Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science 253: 557-560.
- Crameri R, Faith A, Hemmann S, Jaussi R, Ismail C, et al. (1996) Humoral and cell-mediated autoimmunity in allergy to Aspergillus fumigatus. J Exp Med 184: 265-270.
- Pillon R, Ziberna F, Badina L, Ventura A, Longo G, et al. (2015) Prevalence of celiac disease in patients with severe food allergy. Allergy 70:1346-1349.
- Ura B, Scrimin F, Zanconati F, Arrigoni G, Monasta L, et al. (2015) Two-dimensional gel electrophoresis analysis of the leiomyoma interstitial fluid reveals altered protein expression with a possible involvement in pathogenesis. Oncol Rep 33: 2219-2226.
- Fulton KM, Martin SS, Wolfraim L, Twine SM (2013) Methods and applications of serological proteome analysis. Methods Mol Biol 1061: 97-112.
- Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R (2000) Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci, USA, 97: 9390-9395.

