Review Article - (2017) Volume 7, Issue 3
Mohammad Shahid*
Department of Biochemistry and Molecular Biology, College of Medicine, Prince Sattam bin Abdulaziz University, Alkharj, Kingdom of Saudi Arabia, Saudi Arabia
Corresponding Author:
Mohammad Shahid
Department of Biochemistry and Molecular Biology
College of Medicine, Prince Sattam bin Abdulaziz
University, Alkharj, Kingdom of Saudi Arabia
Tel: +966552551527
E-mail: dr.shahid90@yahoo.com
Received Date: February 22, 2017; Accepted Date: June 02, 2017; Published Date: June 10, 2017
Citation: Shahid M. Single Nucleotide Polymorphism (SNPs) in the Genes Associated with Tooth Agenesis. Eur Exp Biol 2017, 7:17. doi: 10.21767/2248-9215.100017
Copyright: © 2017 Shahid M. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Objectives: The main focus of this review article was to collate up to date knowledge with regard to significance of single nucleotide polymorphisms (SNPs) in various genes associated with tooth agenesis. Failure to develop complete set of teeth also called tooth agenesis is a common developmental abnormality manifested mainly as an isolated condition. This anomaly is also associated with many developmental syndromes.
Methods: We reviewed the evidence from the literature with regard to SNPs in many genes associated with this developmental anomaly. The information contained in this review deals only with non-syndromic form of tooth agenesis. This condition generally affects third molars or one or few other permanent teeth, however, in some cases its severity is relatively prevalent.
Results and Conclusions: Mutations in genes such as Msh homeobox 1 (MSX1), Paired box gene 9 (PAX9), Axis inhibitor protein 2 (AXIN2) and Ectodysplasin A (EDA) have been identified that are associated with the familial form of the disease. It has been shown that the phenotypes associated with these mutations indicate the involvement of more complex mechanisms.
Clinical Significance: Evidence collected so far has immense clinical significance to clinical dentists in providing comprehensive guide outlining the role of these gene mutations (SNPs) in various genes and also how the patients affected with this condition will be clinically diagnosed and managed in future.
Keywords
Tooth agenesis; Mutations; Hypodontia; Oligodontia; Polymorphisms
Introduction
Tooth agenesis is genetically and phenotypically a heterogeneous condition. It is assumed that different phenotypic forms are caused by different genes involving different interacting molecular pathways, providing an explanation not only for the wide variety in agenesis patterns but also for associations of dental agenesis with other oral anomalies. Tooth agenesis of one or more teeth comprises one of the most common cranio-facial anomalies in human. It occurs either as an isolated/familial trait or in association with various genetic syndromes. It is transmitted as an autosomal dominant, recessive or X-linked trait. In addition to aesthetic significance, it may affect activities such as mastication, speech alteration and malocclusion. It has been reported in literature that the frequency of missing teeth excluding third molar varies from 2% to 11% in different populations [1]. However, missing wisdom teeth has been reported in 1/5 of the population. Permanent teeth that are found missing include second premolar as most common (3.5%) closely followed by maxillary lateral (1.8%) incisors [2,3]. However, a significant variability with regard to location, symmetry, number of teeth involved, size, shape and developmental stage has been extensively reported in different populations. In most cases, the condition has a genetic basis; however it may also be caused by other factors that include trauma, chemotherapy and radiation therapy and use of thalidomide during pregnancy [4].
Genes Involved in Tooth Development and Differentiation
The molecular aspects of tooth development are similar to those in the development of other organs that include epithelial-mesenchymal interactions. Nearly 300 groups of molecules are involved in tooth development such as fibroblast growth factors (FGFs), bone morphogenetic proteins (BMPs), sonic hedgehog (SHH), and wingless integrated (WNTs). As mentioned previously, BMP4 acts antagonistically with FGF8 and this interaction plays a role in the periodic patterning [5]. The molecular dialogue between oral ectoderm and odontogenic mesenchyme during tooth development is very complicated (Figure 1) [6]. There is a vast knowledge about the signal interchanges that are crucial to control differentiation and morphological changes and spatiotemporal expression of specific genes during teeth formation, yet little is known about their up- or down-regulation [7]. In vitro and in vivo studies have identified that both MSX1 and PAX9 are essential for the induction of bone morphogenetic protein 4 (BMP4), which is considered to be a pivotal step in tooth development in mammals [8]. Evidence so far points to the direction that there is no single gene directly connected with ontogenesis or the lack of any specific tooth. Instead, tooth initiation and morphogenesis occur by an orchestration of numerous genetic and epigenetic factors [6,9]. At the same time, most of the developmental defects in teeth usually occur as a result of mutations in genes encoding signaling molecules and transcription factors [9] such as mutations in the PAX9 gene resulting in partial or total anodontia (missing all teeth, either primary and/or permanent) and mutations in Runt-related transcription factor 2 (RUNX2) causing supernumerary teeth [10-12]. Aim of this review is to present the information associated with the mutations in genes associated with tooth agenesis, therefore, no more information related with the genes involved in tooth development and differentiation is presented in this review.
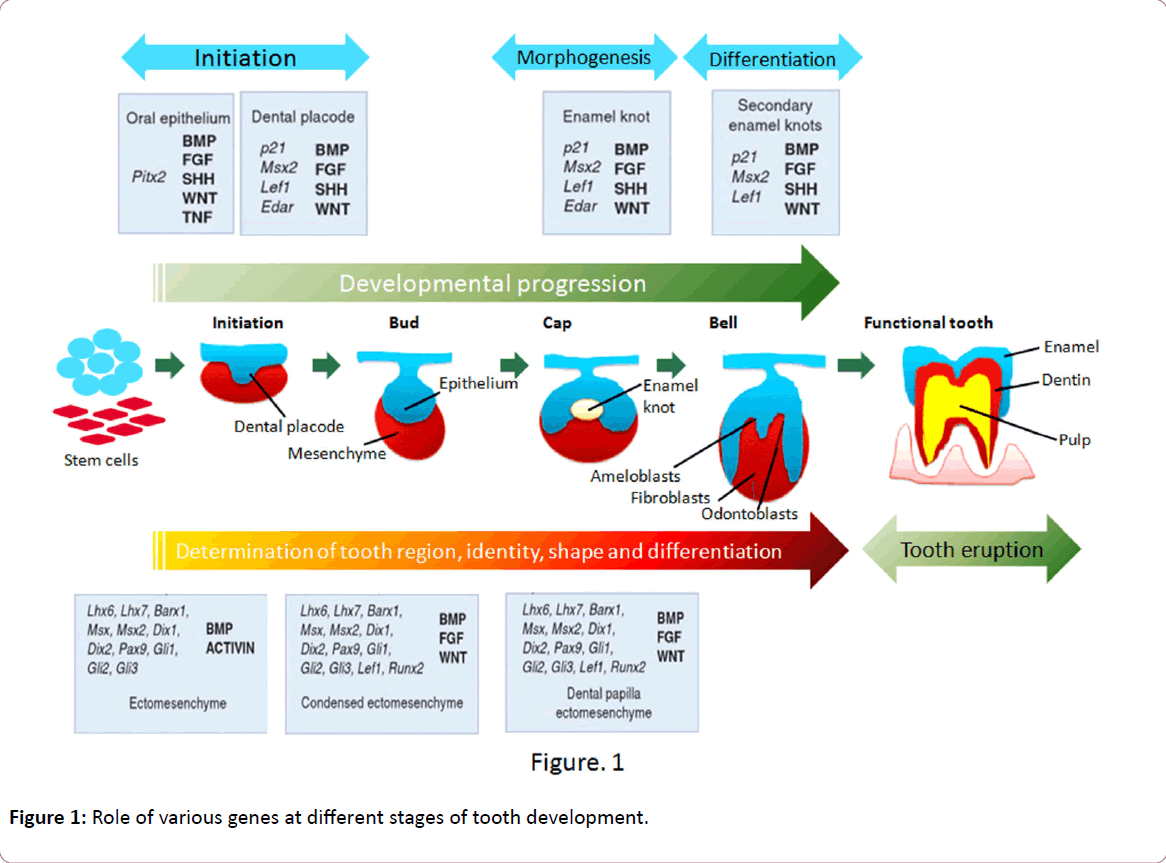
Figure 1: Role of various genes at different stages of tooth development.
The development of tooth is primarily controlled genetically [13]. More than 200 genes have been found to be expressed during this process [14]. Most often reported genes associated with non-syndromic tooth agenesis are PAX9, MSX1, EDA, and AXIN2 [15-20]. Mutations of these genes have an impact on agenesis patterns, although in notably different ways. PAX9 sequence alterations lead to agenesis mainly of molars [15,21] and MSX1 mutations have been implicated primarily in congenitally missing premolars [22,23]. Severe agenesis of both molars and premolars has been noted in patients with an AXIN2 mutation [24] whereas missing incisors are the chief manifestation of EDA-associated non-syndromic oligodontia [20,25-27].
MSX1 Gene
MSX1 gene initially named as homeobox 7 (HOX7) is a nonclustered homeobox protein located on the chromosome 4p16.3-p16.1 [28]. This gene encodes a member of the muscle segment homeobox gene family (Figure 2). The encoded protein functions as a transcriptional repressor during embryogenesis through interactions with components of the core transcription complex and other homeoproteins [29]. It has been shown to have a significant role in tooth development [30]. Mice with a homozygous deletion in Msx1 exhibited a complete secondary cleft palate, complete failure of incisor development, and budstage arrest of molar development [31]. MSX1 was the first gene to be evidently associated with human tooth agenesis [32]. Mutations in this gene have generally been associated with autosomal dominant inheritance of hypodontia (missing up to five permanent teeth, excluding the 3rd molars) as well as oligodontia (missing six or more permanent teeth, excluding 3rd molars or wisdom teeth) [32]. However, in 2006, first association of MSX1 mutations with an autosomal recessive type of hypodontia was reported from Pakistan [17]. Mutations in MSX1 such as p.G22RfsX168 [22], p.M61K [30], p.R176W [33], p.Q187X [16], p.Q189X [34], p.A194V [35], p.R196P [32], p.Q216QfsX125 [33], p.A219T [17], p.A221E [36] has been found to be associated with non-syndromic tooth agenesis. Deletion of chromosome 4p [37] and mutations in MSX1 gene such as p.S104X [38] and p.S202X [39] have also been reported to produce tooth agenesis associated with other symptoms. Recently a novel mutation has been reported in this gene (c. 910_911dupTA; p.*304Tyrext*48) disrupting the original stop codon [40]. Insertion such as 750_751insACCGGCTGCC p.F251Pfsx92 and c.452_9G>A c.451_452insCCCTCAG (7bp ins) p.R151FsX20 [41] have also been recently reported to associated with tooth agenesis. Mutations in MSX1 gene that are associated with tooth agenesis are summed up in Table 1.
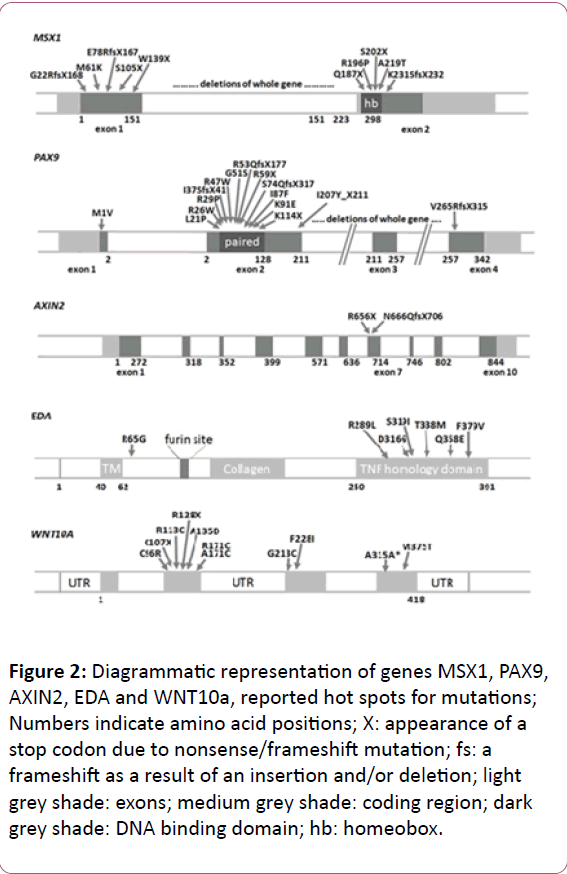
Figure 2: Diagrammatic representation of genes MSX1, PAX9, AXIN2, EDA and WNT10a, reported hot spots for mutations; Numbers indicate amino acid positions; X: appearance of a stop codon due to nonsense/frameshift mutation; fs: a frameshift as a result of an insertion and/or deletion; light grey shade: exons; medium grey shade: coding region; dark grey shade: DNA binding domain; hb: homeobox.
| Phenotype | Mutation | Type | Reference |
|---|---|---|---|
| Oligodontia | *6C>T Polymorphism | Polymorphism | 42 |
| Oligodontia | c.62dupG (p.G22RfsX168) | Frameshift Mutation | 22 |
| Hypodontia | 5354C > G; A40G | Missense mutation | 43 |
| Oligodontia | c.182T>A (p.M61K) | Missense Mutation | 30 |
| Oligodontia | c.314C>A (p.S104X) | Nonsense Mutation | 38 |
| Hypodontia | c.347C>T (G116G) | Missense mutation | 43 |
| Oligodontia | c.416G>A (p.W139X) | Nonsense Mutation | 44 |
| Hypodontia | c.453G>T (p.R151S) | Missense mutation | 45 |
| Oligodontia | c.452_9G>A,c.451_452insCCCTCAG(7bp ins) (p.R151FsX20) | Nonsense mutation | 41 |
| Hypodontia | c.463C>A (p.P155Q) | Missense mutation | 43 |
| Hypodontia | c.517C>A (p.R173S) | Missense mutation | 46 |
| Oligodontia | c.526C>T (p.R176W) | Missense mutation | 33 |
| Oligodontia | c.559C>T (p.Q187X) | Nonsense Mutation | 16 |
| Hypodontia | c.572_573ins GCAAGTT: p.F191fs | Frameshift insertion | 34 |
| Oligodontia | c.581C>T (p.A194V) | Missense Mutation | 35 |
| Oligodontia | c.605C>A (p.S202X) | Nonsense Mutation | 39 |
| Oligodontia | c.614T>G (p.L205R) | Missense mutation | 46 |
| Oligodontia | c.644insA (p.Q216QfsX125) | Frameshift mutation | 33 |
| Oligodontia | c.665G>A (p.A219T) | Missense Mutation | 17 |
| Oligodontia | c.662C>A (p.A221E) | Missense mutation | 36 |
| Oligodontia | c.665_666insA (p.N222KFsX118) | Frameshift Mutation | 47 |
| Hypodontia | c.671 T>C (p.L224P) | Missense Mutation | 48 |
| Oligodontia | c.707delG (p.K237SFsx2), | Frameshift Mutation | 47 |
| Hypodontia | 750_751insACCGGCTGCCp.F251Pfsx92 | Frameshift mutation | 49 |
| Oligodontia | c.910_911dupTA (p.*304Yext*48) | Nonsense Mutation | 40 |
Table 1: Mutations detected in MSX1 gene associated with tooth agenesis.
PAX9 Gene
PAX9 is a member of the paired box (PAX; Figure 2) family of transcription factors and is located on chromosome 14q12-q13 [50]. It plays an integral role both during foetal development and cancer growth [51]. It is evident that this gene also plays important roles during tooth development, as shown by its expression pattern, the phenotype of transgenic mice lacking both copies of the gene, and by the association of agenesis of posterior dentition with PAX9 mutations in humans.10 [52]. Probably it mediates its tooth-specific functions through its interactions with other similar proteins. It has been shown that an important partnership between the PAX9 paired domain protein and the MSX1 homeoprotein is responsible for regulating gene expression in dental Mesenchyme [53].
PAX9 gene was first shown to be associated with all forms of the disease including autosomal dominant, non-syndromic and familial oligodontia [52]. Since then several novel mutations in the gene have been discovered in familial and non-familial forms of the disease. However, a more recent study has found families with affected benign hereditary chorea also showing deletion in PAX9 gene resulting in oligodontia [53]. Mutations reported so far range from being missense that changes just one amino acid in the entire protein to premature stop codons that result in truncation of the protein products. It is also possible that the mutant allele may be present in its hypomorphic state thus indicating the combined activities of both its wild and mutant alleles do not reach the threshold level required for normal tooth development. However, it is also possible that relatively milder phenotypes may result due to defective allele that generates an aberrant protein which acts in a dominantnegative manner or perhaps has a novel function [21,54]. Only a few reports exist in the literature that has been found to be associated with agenesis of deciduous teeth with PAX9 mutations [21,55,56]. Whole gene deletions and the start codon mutations in PAX9 gene caused the most severe teeth developmental defects extending to the premolars and primary molars, i.e. affecting the whole postcanine dentition [55,56]. It also establishes the time and place of morphogenesis [22]. Mutations detected so far in PAX9 gene are summed up in Table 2.
| Phenotype | Mutation | Type | Reference |
|---|---|---|---|
| Oligodontia | c.1A?G (p.M1V) | Missense Mutations | 56 |
| Hypodontia | c.16G?A (p.G6R) | Missense Mutations | 57 |
| Hypodontia | c.43T>A (p.F15I) | Missense Mutation | 58 |
| Hypodontia | c.59delC (p.P20Rfs*65) | Frameshift Mutation | 59 |
| Oligodontia | c.62T?C (p.L21P) | Missense Mutations | 15 |
| Oligodontia | c.76C?T (p.R26W) | Missense Mutations | 60 |
| Oligodontia | (p.L27P) | Missense Mutations | 61 |
| Oligodontia | c.83G?C (p.R28P) | Missense Mutations | 62 |
| Oligodontia | (p.I29T) | Missense Mutations | 61 |
| Oligodontia | c.109-110insG (p.I37Sfs*41) | Frameshift Mutation | 63 |
| Hypodontia | c.128G?A (p.S43K) | Missense Mutations | 57 |
| Oligodontia | c.139C?T (p.R47W) | Missense Mutations | 63,64 |
| Oligodontia | c.140G?C (p.R47P) | Missense Mutation | 47 |
| Oligodontia | c.151G?A (p.G51S) | Missense Mutations | 65 |
| Oligodontia | c.167C?T (p.I56T) | Missense Mutation | 47 |
| Oligodontia | c.175C?T (p.R59*) | Nonsense Mutations | 66 |
| Oligodontia | c.175-176ins288 (p.R59Zfs*177) | Nonsense Mutations | 15 |
| Oligodontia | c.597-599dup (p.D200Sfs*13) | Nonsense Mutations | 52 |
| Oligodontia | c.259A?T (p.I87F) | Missense Mutations | 67 |
| Oligodontia | c.271A?G (p.K91E) | Missense Mutations | 15 |
| Oligodontia | c.321_322insG | Frameshift mutation | 68 |
| Oligodontia | c.340A?T (p.K114*) | Nonsense Mutations | 21 |
| Oligodontia | c.433C?T (p.Q145*) | Nonsense Mutations | 18 |
| Hypodontia | c.73-75 delATC (p.C146T) | Missense mutation | 69 |
| Hypodontia | c.480C>G (p.Y160X) | Nonsense Mutation | 70 |
| Oligodontia | c.503C?G (p.A168G) | Missense mutations | 71 |
| Oligodontia | c.619-621 del ATCins24bp(p.I207Yfs*211) | Frameshift Mutation | 67 |
| Oligodontia | c.718G>C (p.A240P) | Missense Mutation | 72 |
| Oligodontia | c.792-793insC (p.V265Rfs*315) | Frameshift Mutation | 73 |
| Hypodontia | g.718C (C Allele), C912T (T Allele),A1031G (G allele) | Polymorphism | 74 |
| Oligodontia | g.10672A > T (p.D40I) | Missense Mutation | 43 |
Table 2: Mutations in PAX9 gene associated with tooth agenesis.
AXIN2 Gene
AXIN2 or axis inhibitor protein 2 is a gene located on chromosome 17q23-q24 [75]. It is a signaling molecule in the ‘wingless-type MMTV integration site family’ (WNT) pathway that starts operating very early in tooth placode formation (Figure 2). The association of the gene to tooth agenesis was first found in a Finnish family with a predisposition for colorectal cancer [24]. It has been shown that the AXIN2 mutations may also be responsible for sporadic forms of incisor agenesis [76]. Mutations in human AXIN2 associated with oligodontia have been reported [24,33,77]. In humans, mutations in AXIN2 cause tooth agenesis that affects permanent teeth predominantly, including the permanent molars, lower incisors, and upper lateral incisors. AXIN2 is involved in sporadic forms of common incisor agenesis [78,79]. Mutations detected so far in AXIN2 gene are summed up in Table 3. LTBP3 (latent transforming growth factor beta binding protein 3) is a gene that modulates the bioavailability of Transforming growth factor beta 1 (TGF-β) [80,81]. This gene is located on chromosome 11q12 [82]. This gene has been found to be the causal factor in a Pakistani family showing autosomal recessive form of familial oligodontia [83].
| Phenotype | Mutation | Type | Reference |
|---|---|---|---|
| Oligodontia | c.923C>T (p.T308M) | Missense Mutation | 78 |
| Hypodontia | c.1387 C>T (p.R463C) | Missense mutation | 79 |
| Oligodontia | c.C1978T (p.H660Y),c.1966C>T (p.R656Stop) and c.1994delG (p.L688Stop) | Missense mutation Nonsense Mutation |
24 |
| Oligodontia | c.1989G>A (p.W663X) | Nonsense mutation | 80 |
| Oligidontia | c.1999dupG (p.G666GfsX42) c.2051C>T (p.A684V) c.2272G>A (p.A758T) |
Frameshift Mutation Missense mutation Missense mutation |
33 |
| Oligodontia | c.2490G>C (p.M830I) | Missense Mutation | 78 |
Table 3: Mutations found in AXIN2 gene related with tooth agenesis.
EDA1 Gene
EDA1 (ectodysplasin 1) is located on chromosome Xq12-q13.1 (Figure 2) that encodes a protein which belongs to the tumor necrosis factor (TNF) superfamily of ligands. In total, eight isoforms of the EDA transcript can be created by differential splicing of the 12 exons. It has been associated to X-linked recessive form of ectodermal dysplasia [84]. A study of Chinese families with non syndromic X-linked hypodontia showed that a Thr338Met mutation of the EDA gene was responsible for the congenital absence of maxillary and mandibular central incisors, lateral incisors, and canines, with the high possibility of persistence of maxillary and mandibular first permanent molars [26]. Both Ectodysplasin A receptor (EDAR) and Ectodysplasin-A receptor-associated adapter protein (EDARADD) encode respectively the proteins EDAR and EDARADD that belongs to EDA/EDAR/NF-κβ signaling pathway. EDA-EDAR (complex formed by EDA and its receptor) binds to the downstream adaptor EDARADD which leads to activation of the transcription factor NF-κβ (nuclear factor NF-κB pathway). It is essential for the development of hair follicles, teeth, exocrine glands and other ectodermal derivatives [85,86]. Evidence so far also points to the direction that sometime heterozygous mutations in EDA are present in addition to mutations in Wnt Family Member 10A (WNT10A) as digenic mutations [87]. More than seventy mutations in EDA gene have been identified and more than 2/3 of these are reported in conserved TNF homology domain. Only thirteen have been found to be linked with non-syndromic tooth agenesis [88-91]. Of these, five residues (Gln331, Met364, Val365, Ser374 and Ser319) can form a cluster with slightly negative surface potential and play critical role during receptor binding activity of EDA and several other Tumour necrosis factorlike (TNF) domain containing protein [92]. In 2008, a Chinese study reported p.Asp316Gly mutation in EDA protein to be associated with non-syndromic congenital tooth agenesis affecting every tooth type. All affected members show differential expression [93]. Mutations detected so far in EDA and related genes are summed up in Tables 4-6.
| Phenotype | Mutation | Type | Reference |
|---|---|---|---|
| Hypodontia | c.193C>G, (p.R65G) | Missensemutation | 94 |
| Oligodontia | c.252DelT (p.P84PfsX6, c.457C>T(p.R153C), c.466C>T (p.R156C) | Frameshift Mutation Missense mutation |
95 |
| Oligodontia | c.612-29del18bp (IPGIPG205-210del) | Frameshift Mutation | 47 |
| Oligodontia | c.769G>C (p.G257R) | Missense mutation | 95 |
| Oligodontia | c.776C>A (p.A259E) | Missense mutation | 27 |
| Hypodontia | (c.779 T>G) (p.I260S) | Missense mutation | 88 |
| Oligodontia | c.865C>T (p.R289C) | Missense mutation | 27 |
| Oligodontia | c.866G>T (p.R289L) | Missense mutation | 91 |
| Hypodontia | c.874G>T (p.E292X) | Nonsense mutation | 96 |
| Oligodontia | c.901-904delTACT (p.T301SfsX5) | Nonsense mutation | 47 |
| Oligodontia | c.936C>G (p. I312M) | Missense mutation | 95 |
| Hypodontia | c.947A>G (p.D316G) | Missense mutation | 93 |
| Oligodontia | c.956G>T (p.S319I) | Missense mutation | 3 |
| Hypodontia | c.993G>C (p.Q331H) | Missense mutation | 25 |
| Oligodontia | c.1001G>A (p.R334H) | Missense mutation | 27 |
| Hypodontia | c.1013 C>T (p.T338M) | Missense mutation | 93 |
| Oligodontia | c.1045G>A (p.A349T) | Missense mutation | 95 |
| Oligodontia | c.1069C>T (p.R357W) | Missense mutation | 47 |
| Hypodontia | c.1071G>A (p.Q358E) | Missense Mutation | 90 |
| Hypodontia | c.1091T>C (p.M364T) | Missense mutation | 97 |
| Hypodontia | c.1336T>C (p.V365A) | Missense mutation | 89 |
| Oligodontia | c.1133C>T (p.T378M) | Missense mutation | 47 |
| Oligodontia | c.1135T>G (p.F379V) | Missense mutation | 91 |
Table 4: Mutations found in EDA gene associated with tooth agenesis.
| Phenotype | Mutation | Type | Reference |
|---|---|---|---|
| Oligodontia | c.43G>A (p.V15I); c.319A>G (p.M107V), c.871G>A (p.A291T) | Missense Mutations | 95 |
| Oligodontia | c.973C>T (p.R325W); c.1073G>A (p.R358Q) | Missense Mutations | 47 |
| Oligodontia | c.1072C > T (p. R358X) | Nonsense Mutation | 98 |
| Oligodontia | c.1109T>C (p.V370A) | Missense Mutations | 95 |
| Oligodontia | c.1135G>A (p.E379K) | Missense Mutation | 47 |
| Oligodontia | c.1138A>C (p.S380R) | Missense Mutation | 95 |
| Oligodontia | c.1172T>A (p.M391K) | Missense Mutation | 47 |
Table 5: Mutations found in EDAR gene associated with tooth agenesis.
| Phenotype | Mutation | Type | Reference |
|---|---|---|---|
| Oligodontia | c.27G>A (p.M9I) | Missense Mutation | 95 |
| Oligodontia | c.308C>T(p.S103F), c.508C>T (p.R170W) | Missense Mutation | 47 |
Table 6: Mutations found in EDARADD gene associated with tooth agenesis.
WNT10A Gene
WNT10A located adjacent to another gene of the same family Wnt Family Member 6 (WNT6) at 2q35 in humans is active during the development of hair follicles and limbs (Figure 2), and in haematopoiesis. It has been found to be expressed in many adult tissues such as lymph nodes, blood, adrenal gland, prostate, testis, ovary, retina, brain, lung, kidney etc. and may also play an important role in many cancers. Functional studies showed that WNT10A activates the canonical WNT pathway and regulates mesenchymal cell fate in that it inhibits adipogenesis and stimulates osteoblastogenesis [99]. The canonical WNT-β catenin pathway is essential for tooth morphogenesis of both primary and secondary dentition [100]. A recent study in mice has shown that inhibition of the canonical WNT signalling arrests tooth development at early stages [101] whereas constitutive activation of epithelial WNT/β catenin signalling leads to continuous tooth generation [102]. WNT10A is a key mediator of WNT signaling in tooth morphogenesis and, in line with this, WNT10A has been found to be associated with inherited tooth agenesis [103,104]. Van den Boogaard and co-workers have shown that more than half of the patients with non-syndromic oligodontia carry either bi-allelic or mono-allelic mutations in WNT10A [105,106]. In addition to this, WNT10A mutations have also been found in a large proportion of patients having oligodontia and mild manifestations of ectodermal dysplasia.106 WNT/β-catenin signaling has been also reported to play an important role in establishing the size and morphology of the adult tooth [89]. Mutations detected in WNT10a gene are summed up in Table 7.
| Phenotype | Mutation | Type | Reference |
|---|---|---|---|
| Hypodontia | 21-bp deletion combined with 1-bp insertion, c.-14_7delinsC |
Frameshift Mutation | 107 |
| Oligodontia | c.208C>T (p.R70W) | Missense Mutation | 47 |
| Hypodontia | c.321C> A (p.C107*) | Nonsense Mutation | 105 |
| Oligodontia | c.337C>T (p.R113C), c.433G>A (p.V145M), c.433G>T (p.V145L), c.460C>A (p.L154M), c.493G>A (p.G165R) |
Missense Mutation | 47 |
| Oligodontia | c.511C>T (p.R171C) | Missense Mutation | 89,95 |
| Oligodontia | c.579-592del (p.E194AfsX28) GGAACACCCAGCCC |
Frameshift Mutation | 47 |
| Oligodontia | c.637G>A (p.G213S) | Missense Mutation | 95 |
| Oligodontia | c.664G>T (p.E222X) | Nonsense Mutation | 47 |
| Hypodontia | c.682T>A (p.F228I) | Missense Mutation | 47,105 |
Table 7: Mutations in other genes including WNT10A associated with tooth agenesis.
The information available in the literature with regard to reported mutations such as PAX9 (A240P) has been found to be polymorphic in nature at least in sporadic oligodontia in African, American, and European subpopulations. However, a recent study [108] could not establish the presence/absence of a genotype-phenotype association in two Mexican families. The inconsistency of these results might be explained to some extent by population diversities that may cause variation, as genetic polymorphisms often show ethnic disparity. On the other hand, this variation between the high prevalence of tooth agenesis and relatively small number of findings points to the direction that tooth agenesis may be a heterogeneous trait. Combination of two or more gene polymorphisms could result changes in gene structures, gene-gene and protein-protein interactions as well as a reduction in gene dosages, thereby leading to specific phenotypes. Evidence from animal studies suggests that a single homozygous mutation either in MSX1 or PAX9 gene exhibit early arrest of tooth development [51,109]. Contrary to human findings, studies in mice having a heterozygous Pax9 or heterozygous Msx1 loss-of-function mutation do not exhibit tooth developmental defects thus suggesting that different gene dosages are required for tooth development. Recent studies in mice have further strengthened this evidence that synergistic interactions are occurring during the mouse tooth morphogenesis [110]. Further to this; in vitro studies have also provided the evidence that PAX9 expression is down regulated via its interaction with MSX1 during tooth formation [111]. Therefore to further understand and broaden our knowledge about tooth agenesis, we need to perform larger case-control studies.
Conclusion and Future Directions
This review provides further insight to the orthodontists to comprehensively ask the patient about their family history of individuals having similar/dissimilar or any other dental anomalies. Observations with regard to familial and clinical history combined with molecular genetics expertise might bring about tremendous gains for better understanding of genetic causes of various tooth anomalies. Therefore, it is important that both molecular geneticists and dentists must work in tandem to further identify genes causing tooth agenesis. This information will provide enormous information not only about the underlying dental defect but also about the precise changes in the physiologic processes by the mutations in these genes. Identification of mutated genes that cause tooth agenesis (including familial, non-familial, syndromic and non-syndromic) will also enable the future studies to assess the mechanism how environmental and dietary factors modify gene expression and result in distinct but similar clinical phenotypes. Precise exposition of the pathogenic mechanism in tooth agenesis will provide further insights into the role of teeth in craniofacial development and will advance our understanding of cranioorofacial dysmorphology. Evidence gathered through human molecular genetics approach provides information that tooth agenesis is heterogeneous in nature suggesting the involvement of multigenic defects contributing to the varied clinical variability of this entity. Tooth development is a very multifaceted process and involves many genes. This review summarizes the role of mutations in genes such as MSX1, PAX9, AXIN2 etc. in causing tooth agenesis. It is still a long way to go until we finally reach our goal of better understanding the process of ontogenesis. Cross collaboration of geneticists and dentists will be reformed in this century. It is also very important to consider the involvement of epigenetic factors that can reduce gene dosage, alter genes interaction and other posttranscriptional modulation agents that also could explain dental agenesis associated or not with systemic anomalies. Emerging research areas such as human dento-orofacial genetics will become an integral part of the modern health care and will have significant impact the way we diagnose, prevent, and treat these disorders. The identification of mutations in these genes result in mutated/truncated protein and has been linked evidently with tooth agenesis. In addition to other factors, metabolic state of an individual also plays an important role in tooth development. It is anticipated that both environmental and systemic instability will result in the tooth anomalies. Therefore, it becomes quite important that diagnosis of this condition should be made precisely based on detailed account of clinical history, family history and any exposure to environmental factors such as irradiation, use of chemotherapeutic agents and dioxins. Therefore it becomes imperative to find out the actual cause of tooth agenesis in every patient.
It has been demonstrated that mutations in genes such as PAX9, MSX1, and AXIN2 etc. are associated in human tooth agenesis; it is becoming progressively more obvious that many more genes might also play an integral role in tooth development. This is undoubtedly supported by current literature classifying three specific blueprints in human tooth agenesis as: missing pattern of molars and premolars; anterior patterns involving agenesis of cuspids and/or incisors and mixed patterns with missing premolars and lateral incisors [7]. Lack of mutations in PAX9 or MSX1 in families showing a distinct pattern of posterior tooth agenesis further supports the role of so far unidentified genes in causing tooth agenesis [112]. Buccal cells are excellent source of DNA. Dental clinicians during their interactions with patient can easily collect buccal cells noninvasively using tooth brush provided the patients consent to do as well. This non-invasive approach will be more productive as it will increase the patient number ready to take part in molecular genetic studies. Therefore, more and more patients consenting to do this type of analysis will help the molecular geneticists to broaden their knowledge of tooth agenesis. Further studies in this area in different population will be helpful in further understanding the role of genes involved in tooth agenesis. It may presumably show that human dental agenesis is caused by several independent defective genes, acting alone or in combination with other genes and leading to a specific phenotypic pattern. These insights will significantly add to our knowledge of the complex cellular events that give rise to the tooth and of the molecular developmental strategies that control the patterning of the human dentition. In addition further identification of mutations in still undiscovered genes will also shed some more light to better understand this process of odontogenesis. This new knowledge will better equip the clinical dentists to diagnose and treat the patients with this developmental anomaly more accurately. Moreover, this approach will further help to personalize the dental treatment. Further advances to understand the complexity of genetic networks regulating the odontogenesis and how these gene mutations alter gene expression especially in larger cohorts will pave the way for developing an early and accurate diagnostic test to screen the affected families.
Clinical Significance
Many common genetic diseases are not inherited as single gene defect. Gene-environment interactions are important determinants. In the last decade, there is an increasing emphasis toward the evidence-based approach both in medical and dental health practice. Recent scientific advances have improved and impacted on the quality of life of many individuals. Many diseases can now be identified in its initial stages and few are diagnosed even before their onset. Hence it is quite evident that genetics plays an important role in diagnosing and treating these diseases. From dental point of view, both diet and dentists play a significant impact on one’s oral health. The new information will allow dentists to use patient’s medical history to help tailor a dental care plan specific to their needs. If one is at an increased risk of oral disease due to genetic factors, they may need special dental care treatments. People suffering from gum diseases and tooth decay may have genetic variants in some genes. However, sometimes either activation of some genes or altered expression can also result in dental anomalies. Recent developments in dental genetics don’t mean that oral health in under imminent threat. Several genetic factors can influence one’s oral health. These factors can determine the way one’s teeth are aligned and/or whether or not people will get cavities, regardless of their dental care habits. The chemical composition of saliva also determines how well it can neutralize the acids that lead to plaque formation and subsequent tooth decay. Periodontitis is also linked to genetics. Periodontal disease is one of the most common oral diseases characterized by inflammation and destruction of the periodontium. This disease results from complex gene-environment interaction. Multiple gene variants in a particular set of genes appear to confer genetic susceptibility to environment risk factors such as tobacco products, microbial infections, medical and immunological compromise, alcohol consumption, osteoporosis and variety of medications, sex and age. Hence, the treatment of oral, dental, and craniofacial diseases has drastically changed because of these new genomic advances, which are leading the researchers and clinicians to better understand oral biology. More precise and faster diagnostic tests, new drugs and biologics, practicebased research, and culturally sensitive interventions are providing novel avenues to improve oral health.