Research Article - (2017) Volume 3, Issue 3
Hasibe Cingilli Vural*
Department of Medical Biology, Necmettin Erbakan University, Turkey
*Corresponding Author:
Hasibe Cingilli Vural
Department of Medical Biology
Meram Faculty of Medicine
Necmettin Erbakan University, Turkey.
Tel: +90 3322231873
E-mail: hcvural@gmail.com
Received date: July 28, 2017; Accepted date: August 03, 2017; Published date: August 08, 2017
Citation: Vural HC (2017) Tcf7l2 Gene Polymorphism in T2dm with Patients in Turkish Population. J Clin Epigenet. 3:27. doi: 10.21767/2472-1158.100061
Diabetes Mellitus (DM) is a group of metabolic diseases characterized by hyperglycemia resulting from defects of insulin secretion and/or increased cellular resistance to insulin. Chronic hyperglycemia and other metabolic disturbances of DM lead to long-term tissue and organ damage as well as dysfunction involving the eyes, kidneys, and nervous and vascular systems. Type 2 Diabetes mellitus; is the most common form of DM worldwide, and its prevalence is increasing. Its underlying defects can vary from predominant insulin resistance with relative insulin deficiency to a predominant insulin secretory defect with insulin resistance. Transcription 7-like 2 (TCF7L2) protein has been implicated in blood glucose homeostasis. The transcription factor 7-like 2 (TCF7L2) gene is a member of the T-cell factor (TCF)/lymphoid enhancing factor family which encodes a high mobility group (HMG) box-containing transcription factor that plays a key role in the Wnt signaling pathway. The key effector of the canonical Wnt signaling pathway (defined as Wnt pathway hereafter) is the bipartite transcription factor β-cat (β-catenin)/TCF, formed by β-cat and a member of the TCF family [TCF-1/TCF7, LEF-1, TCF-3/TCF7L1 and TCF-4/TCF7L2]. In the absence of Wnt signaling, these HMG box TCF proteins function in the nucleus as transcriptional repressors of the Wnt target genes. TCF7L2 gene is located in chromosome 10q25.3, spans 215.9 kb and contains 14 exons (NCBI build 36.2). However, a previous study has shown that TCF7L2 has 17 exons, of which five are alternative splicing. Genetic variants of gene are associated with increased risk of type 2 diabetes. Common polymorphisms of the transcription factor 7-like 2 gene (TCF7L2) have recently been associated with Type 2 Diabetes all population. The prevalence of Type 2 Diabetes shows wide ethnic and geographic variations.
In this study, we analyzed three polymorphisms in the TCF7L2 gene, rs12255372, rs11196205 and rs7901695 using a case-control design in 163 individuals (113 unrelated T2DM patients and 50 normoglycemic controls) to assess their association with T2DM risk in the Turkish population.
Keywords
Diabetes; TCF7L2 gene; Genetic polymorphism
Introduction
Diabetes mellitus have seen an explosive increase in the number of people diagnosed worldwide in the past two decades. Diabetes mellitus is distinguished into two of which 90% of have Type 2 (non-insulin-dependent), and within this category no more than 10% can be Type 1 Diabetes [1,2]. Thus, most diabetes in the world is be responsible for “common” Type 2 Diabetes. Type 2 Diabetes Mellitus (T2DM) is a complex metabolic disease interplay of genetic and environmental factors [3,4]. The development of diabetes are involved several pathogenic processes which are autoimmune destruction of the β-cells of the pancreas with consequent insulin secretion abnormalities that result in resistance to insulin action [5-7] and caused by alterations in several gene products. Identification of the genes responsible for the pathogenesis of the disease is very important [8,9]. Transcription 7-like 2(TCF7L2) protein has been implicated in blood glucose homeostasis (Figure 1). The transcription factor 7-like 2 (TCF7L2) gene is a member of the T-cell factor (TCF)/lymphoid enhancing factor family which encodes a high mobility group (HMG) box-containing transcription factor that plays a key role in the Wnt signaling pathway [10-12]. Wnt signal pathway is a key component that the regulation of cell growth, cell adhesion and motility, cell polarity, and cell differentiation [13]. Wnt/β-catenin pathway regulates cell differentiation and proliferation through activation of β-catenin/T-cell factor (TCF)- mediated transcriptional activation of Wnt-target genes [14,15]. Wnt signaling activates by the binding of Wnt to their receptor complex, which results in the release of cytosolic β-catenin in the nucleus. β-catenin heterodimerizes with the TCF/lymphoidenhancing factor family of transcription factors to regulate the expression of specific target genes [16-18]. TCF7L2 gene is located in chromosome 10q25.3, spans 215.9 kb and contains 14 exons (NCBI build 36.2). However, a previous study [19] has shown that TCF7L2 has 17 exons, of which five are alternative splicing. Genetic variants of gene are associated with increased risk of Type 2 Diabetes [20,21]. The prevalence of Type 2 Diabetes shows wide ethnic and geographic variations. These polymorphisms reported with Type 2 Diabetes Mellitus in Amish [7], French [8], Indian [9] and other multiple ethnic groups [22]. TCF7L2 polymorphisms in diabetes cases varies between ethnic groups. Therefore, priority should be underlined that the identification of the molecular mechanisms associated with this study. Also a patient with Type 2 diabetes should be evaluated individually, and it is intended to demonstrate the absolute necessity of the test for the presence of gene variants TCF7L2.
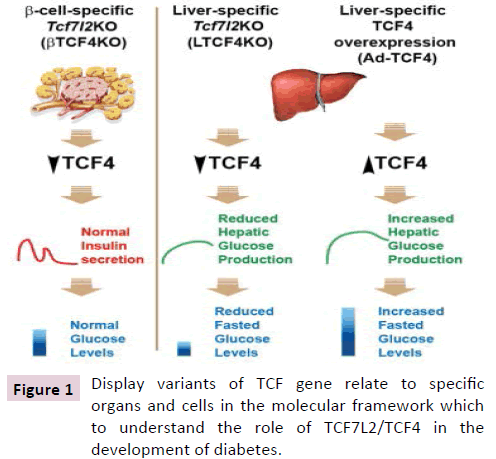
Figure 1: Display variants of TCF gene relate to specific organs and cells in the molecular framework which to understand the role of TCF7L2/TCF4 in the development of diabetes.
Materials and Methods
Study subjects
The study included 113 Type 2 Diabetic patients and 50 normal glucose tolerant control or healthy from Meram Research Hospital in KONYA. The study was conducted in accordance with the Helsinki Declaration. Peripheral blood was drawn from patients and controls and the DNA was extracted from whole blood using the EZ1 Nucleic acid isolation analyser. Briefly, to determination genotyping of rs12255372(G/T), rs11196205(C/G) and rs7901695(C/T) polymorphisms, genomic DNA was isolated from peripheral blood lymphocytes that obtained using EZ1 Nucleic acid isolation analyser (QIAGEN, 2007). All enrolled patients gave written informed consent for participation in the study. The study protocol was approved by the Ethics Committee of the Hospital. No conflict of interest is declared.
Genotyping
We analyzed three polymorphisms in the TCF7L2 gene, rs12255372, rs11196205 and rs7901695 using a case-control design in 163 individuals (113 unrelated T2DM patients and 50 normoglycemic controls) to assess their association with T2DM risk in the Turkish population. Amplification of TCF7L2 gene TCF7L2 gene will be amplified by PCR as previously described. The PCR product will be detected using electrophoresis on 3% agarose gel, The amplified DNA sequence of interest will be purified using Bio Basic purification Kit according to manufacturers instruction. These -rs polymorphisms were genotyped using the PCR based RFLP method. Briefly the region was amplified with following primers illustrated in Table 1.
| rs12255372 | FORWARD, 5'-GGACTTGATTGTTGATTATGGGC-3', REVERSE, 5'-TCTGGCACTCAGAAGAGAGTCAG-3' |
| rs11196205 | FORWARD, 5'-CTGAATCAATGCCATATTGTTGG-3', REVERSE, 5'-ACCATAACTCTCTTACATACAGCT-3' |
| rs7901695 | FORWARD, 5'-CAGATCTGTGCAGTCATCCACAC-3' REVERSE, 5'-CCACTCATAGGACTATACCCAGG-3' |
Table 1: The rs12255372, rs 11196205 and rs7901695 polymorphisms were genotyped using the following primers.
The 200 base pairs PCR product was digested with the TSP509I restriction enzyme (New England Biolabs, USA). The G/T allele creates a restriction site and is gives two fragments 126 and 104 base pairs after digestion with the restriction enzyme (Table 2).
| OMIM Number | Gene | Chromosome position | Location | Polymorhism |
|---|---|---|---|---|
| 602228 | TCF7L2 | 10q25.2– 25.3 | Intron 4 | rs12255372 G/T |
| 602228 | TCF7L2 | 10q25.2– 25.3 | Intron 4 | rs11196205…None |
| 602228 | TCF7L2 | 10q25.2– 25.3 | Intron 3 | rs7901695…..None |
OMIM: Online Mendelian Inheritance in Man
Table 2: Characteristics of the studied polymorphisms in Turk Population.
Statistical Analysis
Statistical analyses were performed using SPSS for Windows software (version 12.0; SPSS, Chicago, IL). The genotype frequencies were tested for Hardy-Weinberg equilibrium using a x2 test.
Results and Discussion
TCF7L2 is implicated in a large variety of diseases. Several single nucleotide polymorphisms are associated with type 2 diabetes. Significant associations were found between the a polymorphism (rs12255372) respectively, the other rs11196205 and rs7901695 in the TCF7L2 gene and Type 2 Diabetes risk, and the other two polymorphisms were not found to be significantly associated with Type 2 Diabetes. All populations analyses show that significant associations are not found between the three SNPs (rs12255372, rs11196205, rs7901695) and the type 2 diabetes in some ethnic populations [23,24]. Briefly, The study proved that the rs12255372 variant of the TCF7L2 gene is associated with Type 2 Diabetes in Turk population. The study confirmed that the genetic variant rs12255372 in the TCF7L2 gene was associated with type 2 diabetes in Turk population (Figures 2 and 3). The results are similar to results observed in other populations [3]. The TCF7L2 gene is an important factor regulating insulin secretion, which could explain its association with Type 2 Diabetes [10].
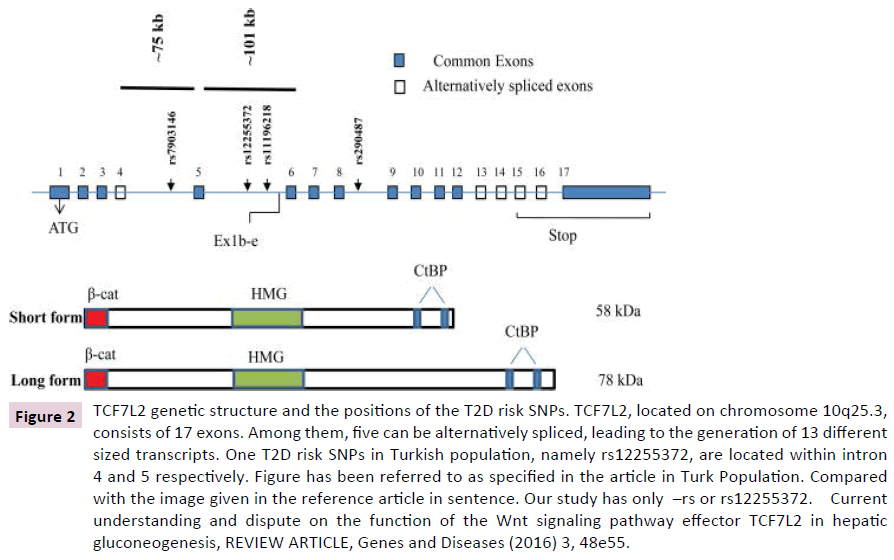
Figure 2: TCF7L2 genetic structure and the positions of the T2D risk SNPs. TCF7L2, located on chromosome 10q25.3, consists of 17 exons. Among them, five can be alternatively spliced, leading to the generation of 13 different sized transcripts. One T2D risk SNPs in Turkish population, namely rs12255372, are located within intron 4 and 5 respectively. Figure has been referred to as specified in the article in Turk Population. Compared with the image given in the reference article in sentence. Our study has only –rs or rs12255372. Current understanding and dispute on the function of the Wnt signaling pathway effector TCF7L2 in hepatic gluconeogenesis, REVIEW ARTICLE, Genes and Diseases (2016) 3, 48e55.
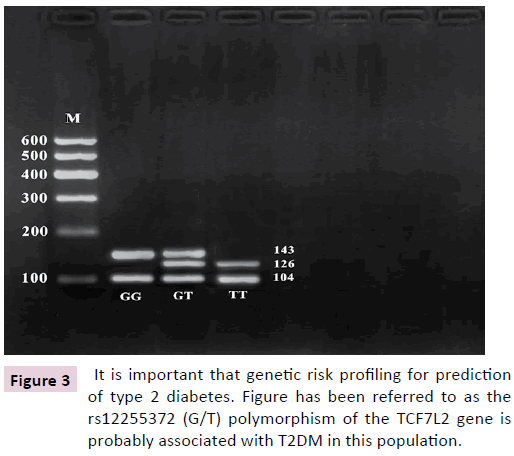
Figure 3: It is important that genetic risk profiling for prediction of type 2 diabetes. Figure has been referred to as the rs12255372 (G/T) polymorphism of the TCF7L2 gene is probably associated with T2DM in this population.
Variants with T2DM, some few have also shown a strong association with the rs12255372 (G/T) variants [8,9]. On the contrary, no association between rs11196205 and rs7901695 variants and T2DM have been found in Turk population.
The relationship between the TCF7L2 variants gene and T2DM has never been studied in Central African populations where T2DM is very prevalent, with high morbidity and mortality rates [11]. Hence we decided to set bases with this pilot study by investigating the association between the TCF7L2 rs12255372 (G/T) polymorphism and T2DM in a Turk population. PCR followed by digestion with TSP509I - 2% agarose gel electrophoresis followed by ethidium bromide staining and UV transilluminator was performed. The expected product sizes are: normal homozygote GG, 143 bp, 104 bp; mutant homozygote TT, 126 bp, 104 bp; and heterozygote GT, 143, 126, and 104 bp, respectively. We used the 100 bp Molecular weight marker as molecular marker and fragments smaller than 100 bp were not visualized.
To test the hypothesis that SNPs in TCF7L2 associated with type 2 diabetes in other studies were also associated with type 2 diabetes in Turkish population, we tested three SNPs in patients and controls. We have observed a trend towards association between only SNPs (rs12255372) type 2 diabetes. No associations were found between the other two SNPs (rs11196205, rs7901695) and type 2 diabetes.Although many studies have been done in different countries regarding the relationship between T2DM and TCF7L2 gene polymorphisms, any study of Turkish population is not available. Common variants in intronic region of TCF7L2 have been identified as strong predictors of T2DM genetic risk. In this study, we analyzed three polymorphisms in the TCF7L2 gene, rs12255372, rs11196205 and rs7901695 using a casecontrol design in 163 individuals (113 unrelated T2D patients and 50 normoglycemic controls) to assess their association with T2D risk in the Turkish population. rs12255372, also known as G>T is a well-studied SNP in the TCF7L2 gene on chromosome 10. In some studies, it has been linked to slight increases in risk for type-2 diabetes, breast cancer and aggressive prostate cancer. It is important that genetic risk profiling for prediction of type 2 diabetes. Furthermore, rs12255372, common variants (rs12255372) in TCF7L2 seem to be associated with an increased risk of diabetes among persons with impaired glucose tolerance. The rs12255372 (G/T) polymorphism of the TCF7L2 gene is probably associated with T2DM in this population. This variant could help to predict the occurrence of T2DM in the Turk population. Our findings should be confirmed by larger study with more accurate genotyping tools. In summary, Although the exact biological mechanism for the association between the TCF7L2 gene and type 2 diabetes risk remains uncertain, these consistent findings indicate that TCF7L2 gene represents an important locus for predicting inherited susceptibility to type 2 diabetes. The result of this study will enhance our knowledge in the field of genetic background of type 2 diabetes mellitus and will provide the clinicians with a new tool for early recognition of susceptible persons which will help in diagnosis of type 2 diabetes mellitus. The data from this study will likely contribute to the understanding the potential roles of these variants across populations; however, further research is required to identify the underlying factors influencing the risk of T2DM in the Turk population.
Acknowledgment
I am gratefully acknowledge the help of my fellowships in data and sample gathering and analysis for providing some samples used in this study. This study was partially supported by Selcuk University Archeometry-Biotechnology Laboratory and Scientific Research Foundation of Selcuk University (BAP) for providing foundation.