Ya-Fang Gao1, Xiao-Li Liu1, Guo-Dong Li1*, Zi-Gang Qian1*, Yong-Hong Zhang2 and Ying-Ying Liu1,3
1Faculty of Traditional Chinese Pharmacy, Yunnan University of Traditional Chinese Medicine, Kunming, P.R. China
2School of Life Sciences, Yunnan Normal University, Kunming, P.R. China
3Yunnan Institute for Food and Drug, Kunming, P.R. China
Corresponding Author:
Guo-Dong Li and Zi-Gang Qian
Faculty of Traditional Chinese Pharmacy
Yunnan University of Traditional Chinese Medicine
Kunming-650500, P.R. China.
E-mail: gammar116@163.com ; qianzig@aliyun.com
Received Date: August 23, 2018; Accepted Date: September 06, 2018; Published Date: September 10, 2018
Citation: Gao YF, Liu XL, Li GD, Qian ZG, Zhang YH, et al. (2018) The Complete Chloroplast Genome of Coptis teeta (Ranunculaceae), An Endangered Plant Species Endemic to the Eastern Himalaya. Biochem Mol Biol J Vol. 4: No.3:19. DOI: 10.21767/2471-8084.100068
Keywords
Coptis teeta Wallich; Chloroplast genome; Endangered species; IUCN; Phylogenetic
Introduction
Coptis teeta Wallich, a perennial herb of Ranunculaceae, was endemic to Eastern Himalaya with narrow distribution range. It is a shade-tolerant species, mainly distributed in the moist temperate, evergreen, broad-leaved forests in northwest Yunnan, China, and northeast India and it occupied highly specialized niches in temperate oak - rhododendron forests and restricted to elevations between 2350 and 3100 m [1,2]. The rhizome of this species, known as Yunnan goldthread (Yunlian in Chinese), is important Chinese herbology since the period of Sheng-Nong (3000 B.C.) [1]. It has excellent pharmacological activity and was used to treat various diseases such as diarrhea, disorder of glucose metabolism, hypertension, cardiovascular and cerebral vessel diseases [3]. The previous study revealed that the species have highly specific microsite requirements that cannot be met in other habitats. Owing to the over-exploitation, several anthropogenic factors, and environmental disruption, the wild population of C. teeta decreased rapidly in recent years [4]. C. teeta has been listed in IUCN Red List of Threatened Species (https://www.iucnredlist.org/) as endangered species with status “A2cd”. And it is also included in Category II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) [5]. Therefore, it is necessary to protect this endangered plant for its highly economic and ecological values, and for the conservation of biodiversity.
To date, few studies of this species have been performed due to lack of genomic data of C. teeta. The previous studies mainly focus on phylogenetic analysis and biogeographic pattern of Coptis by using two plastid and one nuclear markers including psbA-trnH, trnL-trnF and ITS, and six markers, including five plastid and one nuclear markers, respectively [6,7]. In this study, as a part of the genome sequencing project of C. teeta, we assemble and annotate its complete plastid genome and describing its characteristics.
Materials and Methods
Plant material and DNA extraction
Fresh leaves of C. teeta were collected from Gongshan County (27°73′E, 98°66′N), Yunnan province and voucher specimens were deposited in Yunnan University of Traditional Chinese Medicine. Total genomic DNA was extracted using the modified plant genome kit (Bioteke, Beijing, China). DNA quality was detected by electrophoresis on 1% agarose gel (Figure 1) and 1 μL of DNA sample to test concentration using to the NanoDrop spectrophotometers (ThermoFisher Scientific, Wilmington, Delaware, USA), the result showed that its value is 62.6 ng/μL>50 ng/μL.
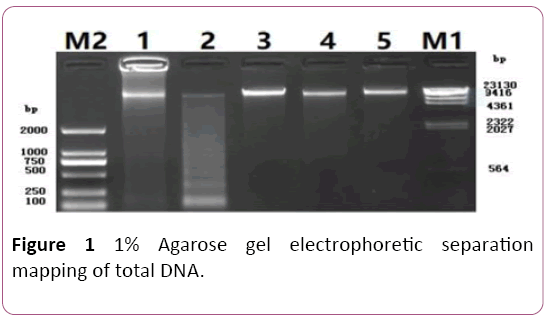
Figure 1: 1% Agarose gel electrophoretic separation mapping of total DNA.
Genome sequencing, assembly and annotation
A sequence library was constructed and sequencing was performed using the Illumina HiSeq 2500-PE150 platform (Illumina, CA, USA). All raw reads were filtered by using NGSQC Toolkit_v2.3.3 with default parameters to obtain clean reads that has discard low quality regions [8]. The plastome was de novo assembled using GetOrganelle pipeline (https:// github.com/Kinggerm/GetOrganelle). The complete chloroplast genome was annotated with the online annotation tool GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) [9], using the published cp genome of C. chinensis (NCBI accession number: NC036485) as a reference sequence, then manual correction was performed with Geneious R11software [10]. The plastid genome map was drawn using OGDRAW program (https:// ogdraw.mpimp-golm.mpg.de/) [11]. The annotated cp genome of C. teeta has been deposited into GenBank with the Accession Number MH359096.
Repeats and simple sequence repeats (SSRs) analysis
REPuter [12] was used to find forward and reversed tandem repeats≥15 bp with minimum alignment score and maximum period size at 100 and 500, respectively. IMEx [13] was used to visualize the SSRs with the minimum repeat numbers set to 10, 5, 4, 3, 3 and 3 for mono-, di-, tri-, tetra-, penta- and hexanucleotides, respectively.
Phylogenetic analysis
The phylogenetic analysis was conducted based on 31 published chloroplast genomes to infer phylogenetic position of C. teeta within the family of Ranunculaceae. The cp genome of Nandina domestica (GenBank: DQ923117) was included as outgroup. The LSC, SSC and one IR region of the total 32 chloroplast genomes were aligned using MAFFT 7.308 [14]. The maximum likelihood (ML) tree was reconstructed by RAxML 8.2.11 [15] with the nucleotide substitution model of GTR+G and node support was estimated by means of bootstrap analysis with 1000 replicates.
Results and Discussion
Characteristics of chloroplast genome of C. teeta
The complete chloroplast genome of C. teeta is a circular DNA with 154,280 bp in length, comprising four subunits: one large singe copy (LSC) (87,519 bp), one small singe copy (SSC) (17,595 bp) and two inverted repeat regions (IRs) (24,583 bp for each) (Figure 2). The overall GC content was 38.3 %. The IR regions had a higher GC content (43.3%) than LSC (36.7%) and SSC regions (32.2%). That was caused by the high GC content of the four ribosomal RNA (rRNA) genes (55.5%) presented in the IR regions, similar to that of C. chinensis Franchet [16].
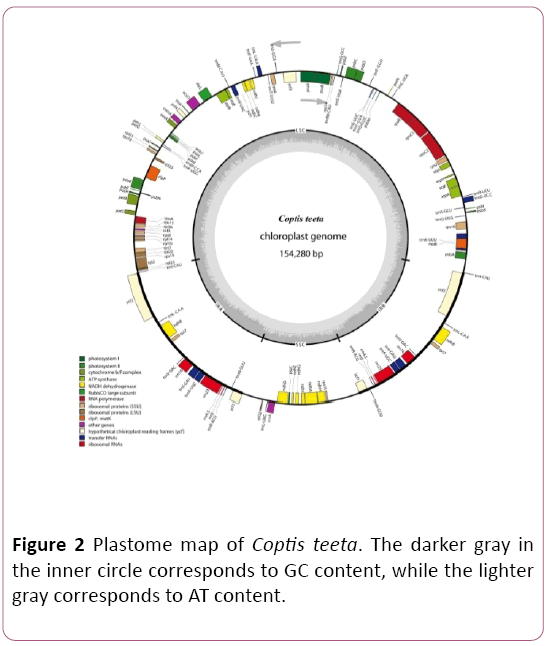
Figure 2: Plastome map of Coptis teeta. The darker gray in the inner circle corresponds to GC content, while the lighter gray corresponds to AT content.
The chloroplast genome of C. teeta contains 125 genes, comprising 81 protein-coding genes (PCGs), 36 transfer RNA (tRNA) genes and 8 rRNA genes. Among these genes, ndhA, ndhB, rpl2, rpoc1, atpF, rps16, trnA-UGC, trnI-GAU, trnV-UAC, trnL-UAA, trnG-UCC, trnK-UUU contain one intron, while the clpP and ycf3 genes contain two introns. The trnK-UUU gene has a larger intron of 2,853 bp compared with other introns. The IR regions include seven tRNAs (trnN-GUU, trnR-ACG, trnA-UGC, trnL-GAU, trnV-GAC, trnL-CAA, trnI-CAU ), four rRNAs without intron (rrn16, rrn23, rrn4.5 and rrn5) and four PCGs (rcf1, rcf2, rps7, ndhB, rps15) and all of these genes are totally duplicated. Additionally, one tRNA (trnL-UAG) and ten PGGs (rpl32, rps15, ccsA, ndhD, Psac, ndhE, ndhG, ndhI, ndhA, ndhH) are contained in SSC region of C. teeta chloroplast genome.
Repeat and SSR analysis
For repeat structure analysis, 42 forward and 23 reverted repeats with minimal repeat size of 15 bp were detected in cp genome of C. teeta (Table 1). Most of these repeats were between 15 and 20 bp. The longest forward repeats were of 39 bp, one sequence of which located in the intergenic region between trnV-GAC and rps7 of inverted repeated regions (IR), the other sequence located in ycf3 of LSC. There are 31 repeats with two sequences started in the same region. Among them, 21 repeats located in the LSC region, 7 located in the IR regions, and 3 located in SSC region. Other 34 repeats with two sequences started in separated regions.
| ID |
Repeat Start 1 |
Type |
Size (bp) |
Repeat Start 2 |
E-Value |
Region |
Gene |
| 1 |
161 |
F |
15 |
94911 |
6.23 |
IRb/LSC |
ycf1; IGS |
| 2 |
1319 |
F |
15 |
146257 |
6.23 |
IRb/SSC |
ycf1; ndhA |
| 3 |
1686 |
F |
18 |
8903 |
0.0974 |
IRb |
trnN-GUU; trnl-GAU |
| 4 |
1729 |
F |
15 |
22897 |
6.23 |
IRb |
trnN-GUU; ycf2 |
| 5 |
2352 |
F |
15 |
34195 |
6.23 |
IRb/LSC |
trnR-ACG; trnS-GCU |
| 6 |
2352 |
F |
15 |
61944 |
6.23 |
IRb/LSC |
trnR-ACG; trnS-UGA |
| 7 |
3964 |
F |
15 |
7706 |
6.23 |
IRb |
rrn23; trnl-GAU |
| 8 |
6084 |
F |
15 |
91596 |
6.23 |
IRb/LSC |
IGS |
| 9 |
6598 |
F |
19 |
7684 |
0.0244 |
IRb |
trnA-UGC; trnl-GAU |
| 10 |
7003 |
F |
15 |
95317 |
6.23 |
IRb/LSC |
trnA-UGC;IGS |
| 11 |
7017 |
F |
18 |
74997 |
0.0244 |
IRb/LSC |
trnA-UGC; trnF-GAA |
| 12 |
8184 |
F |
18 |
8215 |
0.0974 |
IRb |
IGS |
| 13 |
9475 |
F |
15 |
148599 |
6.23 |
IRb/SSC |
rrn16; ndhH |
| 14 |
10179 |
F |
17 |
57871 |
0.39 |
IRb/LSC |
trnV-GAC; trnT-GGU |
| 15 |
11638 |
F |
16 |
38957 |
1.56 |
IRb/LSC |
IGS |
| 16 |
11991 |
F |
39 |
70172 |
2.21E-14 |
IRb/LSC |
IGS; ycf3 |
| 17 |
19923 |
F |
16 |
20059 |
1.56 |
IRb |
ycf2 |
| 18 |
20113 |
F |
16 |
71145 |
1.56 |
IRb/LSC |
ycf2; IGS |
| 19 |
24047 |
F |
16 |
94901 |
1.56 |
IRb/LSC |
ycf2; IGS |
| 20 |
30432 |
F |
20 |
62669 |
0.00609 |
LSC |
IGS |
| 21 |
32229 |
F |
16 |
81798 |
1.56 |
LSC |
IGS |
| 22 |
34192 |
F |
21 |
61941 |
0.00152 |
LSC |
trnS-GCU; trnS-UGA |
| 23 |
35625 |
F |
19 |
62985 |
0.0244 |
LSC |
trnG-GCC; trnG-UCC |
| 24 |
38000 |
F |
16 |
38051 |
1.56 |
LSC |
atpF |
| 25 |
39908 |
F |
17 |
101565 |
0.39 |
LSC |
IGS; petB |
| 26 |
45078 |
F |
16 |
94306 |
1.56 |
LSC |
rpoC2; IGS |
| 27 |
46933 |
F |
16 |
98952 |
1.56 |
LSC |
rpoC1; psbB |
| 28 |
54520 |
F |
16 |
102145 |
1.56 |
LSC |
IGS; petB |
| 29 |
55783 |
F |
16 |
77512 |
1.56 |
LSC |
IGS |
| 30 |
57577 |
F |
17 |
139324 |
0.39 |
LSC/SSC |
IGS |
| 31 |
58768 |
F |
16 |
92720 |
1.56 |
LSC |
IGS |
| 32 |
62639 |
F |
17 |
71507 |
0.39 |
LSC |
IGS |
| 33 |
62772 |
F |
16 |
152783 |
1.56 |
LSC/SSC |
IGS |
| 34 |
63190 |
F |
21 |
92511 |
0.00152 |
LSC |
trnfM-CAU; trnP-UGG |
| 35 |
65213 |
F |
21 |
67437 |
0.00152 |
LSC |
psaB; psaA |
| 36 |
70175 |
F |
37 |
146807 |
3.54E-13 |
LSC/SSC |
ycf3; ndhA |
| 37 |
77524 |
F |
16 |
95015 |
1.56 |
LSC |
IGS |
| 38 |
106539 |
F |
16 |
139969 |
1.56 |
LSC/SSC |
rpl14; IGS |
| 39 |
108087 |
F |
16 |
138701 |
1.56 |
LSC/SSC |
IGS |
| 40 |
138345 |
F |
16 |
153380 |
1.56 |
SSC |
IGS |
| 41 |
139327 |
F |
16 |
150849 |
1.56 |
SSC |
IGS |
| 42 |
139331 |
F |
17 |
153983 |
0.39 |
SSC |
IGS |
| 43 |
412 |
R |
15 |
52362 |
6.23 |
SSC/LSC |
IGS; petB |
| 44 |
1320 |
R |
15 |
17771 |
6.23 |
SSC/IRa |
IGS; ycf1 |
| 45 |
10547 |
R |
15 |
19893 |
6.23 |
SSC/IRa |
psaC; IGS |
| 46 |
11639 |
R |
15 |
101219 |
6.23 |
SSC/LSC |
ndhD; IGS |
| 47 |
12190 |
R |
15 |
118155 |
6.23 |
SSC/LSC |
ndhD; atpA |
| 48 |
30692 |
R |
16 |
74169 |
1.56 |
IRa/LSC |
rps7; atpB |
| 49 |
32228 |
R |
17 |
77513 |
0.39 |
IRa/LSC |
ndhB; ndhK |
| 50 |
38958 |
R |
18 |
101216 |
0.0974 |
IRa/LSC |
ycf2; IGS |
| 51 |
39215 |
R |
16 |
81430 |
1.56 |
IRa/LSC |
ycf2; rps14 |
| 52 |
39633 |
R |
16 |
90680 |
1.56 |
IRa/LSC |
ycf2; rps14 |
| 53 |
40216 |
R |
16 |
62753 |
1.56 |
IRa/LSC |
ycf2; petL |
| 54 |
44548 |
R |
17 |
139326 |
0.39 |
LSC/IRb |
rpl22; ndhB |
| 55 |
44548 |
R |
16 |
150849 |
1.56 |
LSC/IRb |
rpl22; rrn23 |
| 56 |
55782 |
R |
17 |
81799 |
0.39 |
LSC |
psbB; rps4 |
| 57 |
55869 |
R |
17 |
73441 |
0.39 |
LSC |
psbB; atpB |
| 58 |
71197 |
R |
16 |
147968 |
1.56 |
LSC/IRb |
rbcL; trnA-UGC |
| 59 |
73193 |
R |
16 |
108492 |
1.56 |
LSC |
atpB; rpoC2 |
| 60 |
77510 |
R |
16 |
93741 |
1.56 |
LSC |
ndhK; psbC |
| 61 |
77512 |
R |
17 |
81798 |
0.39 |
LSC |
ndhK; rps4 |
| 62 |
81801 |
R |
16 |
93738 |
1.56 |
LSC |
rps4; psbC; |
| 63 |
83628 |
R |
16 |
139655 |
1.56 |
LSC/IRb |
ycf1; ndhB |
| 64 |
85488 |
R |
16 |
149157 |
1.56 |
SSC/IRb |
IGS; rrn23 |
| 65 |
139356 |
R |
16 |
139960 |
1.56 |
IRb |
ndhB |
| |
|
|
|
|
|
|
|
| F: Forward; R: Reverted; IGS: intergenic space |
Table 1: Repeat sequences in C. teeta chloroplast genome.
cpSSRs markers are widely used to study the population genetics and evolutionary processes of wild plants [17,18]. There were totally 62 SSRs in cp genome of C. teeta, most of which were in LSC (Table 2). Among them, 31 (50.0%) were mononucleotide SSRs, fifteen (24.2%) were dinucleotide SSRs, six (9.7%) were tri-nucleotide SSRs, eight (12.9%) were tetranucleotide SSRs, one (0.2%) was penta-nucleotide SSR, and one (0.2%) was hexa-nucleotide SSRs. Only twelve SSRs were located in genes and the others were in intergenic regions. 30 (96.8%) of the mononucleotide SSRs belonged to the A/T type, which were consistent with the hypothesis that cpSSRs were generally composed of short polyadenine (poly A) or polythymine (poly T) repeats and rarely contained tandem guanine (G) or cytosine (C) repeats. These cpSSR markers can be used in the conservation genetics of C. teeta.
| ID |
Repeat Motif |
Length (bp) |
Start |
End |
Region |
Gene |
| 1 |
(T) 10 |
10 |
2610 |
2619 |
IRb |
|
| 2 |
(AATA) 3 |
12 |
11645 |
11656 |
IRb |
|
| 3 |
(ATCT) 3 |
12 |
29571 |
29582 |
LSC |
trnK-UUU |
| 4 |
(A) 10 |
10 |
30435 |
30444 |
LSC |
|
| 5 |
(C) 11 |
11 |
31241 |
31251 |
LSC |
rps16 |
| 6 |
(AT) 7 |
14 |
32232 |
32245 |
LSC |
|
| 7 |
(A) 11 |
11 |
33582 |
33592 |
LSC |
|
| 8 |
(T) 11 |
11 |
33825 |
33835 |
LSC |
|
| 9 |
(T) 10 |
10 |
35187 |
35196 |
LSC |
trnG-UCC |
| 10 |
(T) 10 |
10 |
35734 |
35743 |
LSC |
|
| 11 |
(CTGT) 3 |
12 |
36989 |
37000 |
LSC |
atpA |
| 12 |
(T) 10 |
10 |
40322 |
40331 |
LSC |
|
| 13 |
(T) 10 |
10 |
42352 |
42361 |
LSC |
|
| 14 |
(T) 14 |
14 |
44550 |
44563 |
LSC |
rpoC2 |
| 15 |
(AT) 5 |
10 |
45302 |
45311 |
LSC |
rpoC2 |
| 16 |
(TA) 5 |
10 |
45923 |
45932 |
LSC |
rpoC2 |
| 17 |
(TA) 5 |
10 |
53640 |
53649 |
LSC |
|
| 18 |
(A) 10 |
10 |
54985 |
54994 |
LSC |
|
| 19 |
(TA) 7 |
14 |
55785 |
55798 |
LSC |
|
| 20 |
(TA) 5 |
10 |
55828 |
55837 |
LSC |
|
| 21 |
(T) 13 |
13 |
57582 |
57594 |
LSC |
|
| 22 |
(TTATA) 3 |
15 |
58077 |
58091 |
LSC |
|
| 23 |
(TA) 6 |
12 |
58573 |
58584 |
LSC |
|
| 24 |
(AAAG) 3 |
12 |
59161 |
59172 |
LSC |
|
| 25 |
(A) 10 |
10 |
62672 |
62681 |
LSC |
|
| 26 |
(ATA) 4 |
12 |
68971 |
68982 |
LSC |
|
| 27 |
(A) 11 |
11 |
72877 |
72887 |
LSC |
|
| 28 |
(A) 14 |
14 |
73196 |
73209 |
LSC |
|
| 29 |
(AT) 5 |
10 |
73741 |
73750 |
LSC |
|
| 30 |
(T) 10 |
10 |
75418 |
75427 |
LSC |
|
| 31 |
(TA) 7 |
14 |
77514 |
77527 |
LSC |
|
| 32 |
(ATA) 4 |
12 |
77525 |
77536 |
LSC |
|
| 33 |
(ACCA) 3 |
12 |
78836 |
78847 |
LSC |
trnV-UAC |
| 34 |
(ATA) 4 |
12 |
81391 |
81402 |
LSC |
atpB |
| 35 |
(AT) 7 |
14 |
81801 |
81814 |
LSC |
|
| 36 |
(AT) 6 |
12 |
86612 |
86623 |
LSC |
|
| 37 |
(T) 11 |
11 |
86623 |
86633 |
LSC |
|
| 38 |
(A) 12 |
12 |
86710 |
86721 |
LSC |
|
| 39 |
(T) 10 |
10 |
89930 |
89939 |
LSC |
|
| 40 |
(TA) 6 |
12 |
93742 |
93753 |
LSC |
|
| 41 |
(T) 10 |
10 |
94315 |
94324 |
LSC |
|
| 42 |
(ATA) 4 |
12 |
95016 |
95027 |
LSC |
|
| 43 |
(T) 10 |
10 |
97412 |
97421 |
LSC |
clpP |
| 44 |
(ATT) 4 |
12 |
100305 |
100316 |
LSC |
psbN |
| 45 |
(TTCT) 3 |
12 |
108128 |
108139 |
LSC |
|
| 46 |
(A) 16 |
16 |
108491 |
108506 |
LSC |
|
| 47 |
(TTAT) 3 |
12 |
125025 |
125036 |
IRa |
|
| 48 |
(A) 10 |
10 |
134063 |
134072 |
IRa |
|
| 49 |
(T) 14 |
14 |
139329 |
139342 |
SSC |
|
| 50 |
(AT) 5 |
10 |
139695 |
139704 |
SSC |
|
| 51 |
(AT) 5 |
10 |
143541 |
143550 |
SSC |
|
| 52 |
(AT) 5 |
10 |
145476 |
145485 |
SSC |
|
| 53 |
(ATA) 4 |
12 |
146996 |
147007 |
SSC |
ndhA |
| 54 |
(T) 14 |
14 |
150851 |
150864 |
SSC |
|
| 55 |
(T) 11 |
11 |
151012 |
151022 |
SSC |
|
| 56 |
(CATT) 3 |
12 |
151870 |
151881 |
SSC |
|
| 57 |
(T) 12 |
12 |
152189 |
152200 |
SSC |
|
| 58 |
(T) 11 |
11 |
152711 |
152721 |
SSC |
|
| 59 |
(CTTTTA) 3 |
18 |
152750 |
152767 |
SSC |
|
| 60 |
(A) 11 |
11 |
153416 |
153426 |
SSC |
|
| 61 |
(T) 11 |
11 |
153836 |
153846 |
SSC |
|
| 62 |
(T) 11 |
11 |
153984 |
153994 |
SSC |
|
Table 2: Simple sequence repeats (SSRs) in the C. teeta chloroplast genome.
Phylogenetic analysis
The phylogenetic tree showed that species from the Ranunculaceae formed a monophyletic clade (Figure 3) and the intra-family topology was consistent with previous studies [6,16,19]. The result strongly supported C. teeta and its congeneric species, C. chinensis, as sister group with 100% bootstrap value. This newly reported chloroplast genome will provide new insights into phylogenetic studies within the Ranunculaceae and facilitate future conservation of C. teeta.
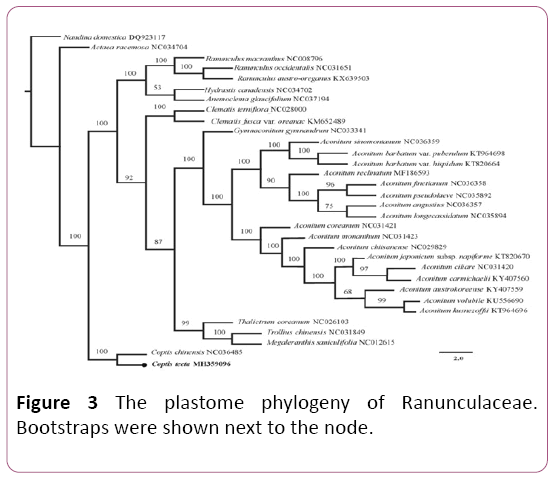
Figure 3: The plastome phylogeny of Ranunculaceae. Bootstraps were shown next to the node.
Conclusion
In this study, we reported and analyzed the first complete chloroplast genome of C. teeta, which are an endemic and endangered plant and a source for famous traditional Chinese medicine.
The circular chloroplast genome exhibits typical quadripartite regions with 154,280 bp in size, including two inverted repeat (IR, 24,583 bp) regions, one large singe copy region (LSC) and one small singe copy region (SSC) of 87,519 bp and 17,595 bp, respectively.
The cp genome of C. teeta was rich in SSRs, which will be informative sources for developing new molecular markers to evaluate genetic diversity and provide effective strategies for conservation of this species. The phylogenetic analysis showed that C. teeta and C. chinensis form one clade as sister group. This information will be useful on phylogenetic analysis of genus Coptis and will also enhance our understanding on the evolutionary relationships among Ranunculaceae.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81560613), Special subsidies for public health services of TCM “the national survey of TCM resources” (DSS, MOF. No 66/2017), and the Key laboratory training program in Yunnan (2017DG006).
References
- Huang J, Long C (2007) Coptis teeta-based agroforestry system and its conservation potential: A case study from Northwest Yunnan. AMBIO 36: 344.
- Pandit MK, Babu CR (1999) Synaptic mutation associated with gametic sterility and population divergence in Coptis teeta (Ranunculaceae). Bot J Linn Soc 133: 526.
- Wang WQ (2016) A review on pharmacologic effects of effective ingredients in Huanglian. Clin J Chinese Med 26: 147-148.
- Pandit MK, Babu CR (1998) Biology and conservation of Coptis teeta Wall: An endemic and endangered medicinal herb of Eastern Himalaya. Envir Conserv 25: 262.
- UNEP-WCMC (Comps) (2014) Checklist of CITES species. CITES Secretariat, Geneva, Switzerland, and UNEP-WCMC, Cambridge, United Kingdom.
- Xiang KL, Wu SD, Yu SX, Liu Y, Florian J, et al. (2016) The first comprehensive phylogeny of Coptis (Ranunculaceae) and its implications for character evolution and classification. PLoS ONE 11: e0153127.
- Xiang KL, Andrey SE, Xiang XG, Florian J, Wang W (2018) Biogeography of Coptis salisb. (Ranunculales, Ranunculaceae,Coptidoideae), an Eastern Asian and North American genus. BMC Evol Biol 18: 74.
- Patel RK, Jain M (2012) NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PloS ONE 7: e30619.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, et al. (2017) GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45: W6-W11.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647-1649.
- Lohse M, Drechsel O, Kahlau S, Bock R (2013) Organellar Genome-DRAW a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41: 575-580.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, et al. (2001) REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29: 4633-4642.
- Mudunuri SB, Nagarajaram HA (2007) IMEx: Imperfect microsatellite extractor. Bioinformatics 23: 1181-1187.
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30: 772-780.
- Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57: 758-771.
- He Y, Xiao HT, Deng C, Fan G, Qin SS, et al. (2017) Complete chloroplast genome sequence of Coptis chinensis Franch and its evolutionary history. BioMed Res Int 1-7.
- Provan J (2009) Novel chloroplast microsatellites reveal cytoplasmic variation in Arabidopsis thaliana. Mol Ecol 9: 2183-2185.
- Flannery ML, Mitchell FJ, Coyne S (2006) Plastid genome characterization in Brassica and Brassicaceae using a new set of nine SSRs. Theor Appl Genet 113: 1221-1231.
- Liu HJ, Xie L (2016) Advances in molecular phylogenetics of Ranunculaceae. Acta Bot Boreali-Occidentalia Sinica 36: 1916-191.




