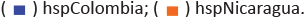Keywords
Evolution; Colombian populations; Gastric cancer; H. pylori; Phylogenomics
Introduction
The Helicobacter pylori (H. pylori) is a Gram-negative bacterium that has colonized half of the world’s population and which induces a chronic inflammatory process [1,2]. H. pylori is the main factor in the development of gastric cancer, which was classified as a type I carcinogen by World Health Organization (WHO) in 1994 [3]. The infection by H. pylori in the human host occurred 88,000-116,000 years ago [4], and has coevolved with human beings since the first migration from East Africa approximately 60,000 years ago [5].
The high genomic diversity of H. pylori is a product of intra and intergenomic mixing process from multiple strains that provides
its adaptation and colonization skills in human populations [6]. Its infection, adaptation, and survival mechanisms are diverse, and the vacA gene induces apoptosis, increased permeability in gastric cell, and causes inhibition of the immune response of T cells [7]. This gene shows high genetic diversity, presenting two allele families, s1/s2 and m1/m2, which are associated with the development of gastric cancer [8]. The AlpA adhesin is constitutive of H. pylori this has as main function the adhesion to gastric epithelium [9]. Also, this gene could induce the IL-6 and IL-8 expression in human host [10].
The incidence of gastric cancer in Latin America has been characterized by being high in the mountains and low on the coasts [11]. In Colombia, in the Andean zone in the city of Tuquerres, the incidence rate is 150/100,000 inhabitants, while on the Pacific coast, in the city of Tumaco it is 6/100,000 inhabitants, in spite of a similar incidence of H. pylori (~90%) [12,13]. Likewise, human populations have different ancestry: in the Andean zone 67% is Native American, 31% is European, and 2% is African. On the other hand, the ancestry of the people in Tumaco is 58% African, 23% Native American, and 19% European [14]. This phenomenon is known as the “Colombian enigma” [12].
Multilocus Sequence Typing (MLST) is a technique that has been used to estimate the evolutionary relations among different strains and to study historical migrations of H. pylori and its host. Some studies that used the MLST technique on H. pylori based on seven housekeeping genes have been able to identify various population groups: hpEurope, hpNEAfrica, hpWAfrica, hpAfrica1, hpAfrica2, hpAsia2, hpSahul, and hpEastAsia. Bacteria subpopulations have been identified: hspAmerindian, hspEAsia, and hspMaori [15-18].
In previous studies it is being found that bacteria that infected human population from American Continent and Colombia in particular belong to European origin (hpEurope) being an important risk factor due to the human host-bacteria evolutive desynchronization [19,20]. However, H. pylori whole genome recent studies have shown the emergence of new independent linages for several countries from Latin America [21-23]. One of the characteristics of H. pylori is its great genetic diversity, and it is known that different ancestry strains might interact differently with their human host clearly influencing pathogenesis [24]. Therefore, to describe the evolutive process of H. pylori could allow us to obtain information about the risk of cancer development. To develop our main aim of this study, we suggested carrying out an analysis of the evolutionary relations of H. pylori from the department of Narino, Colombia, who had a different risk of gastric cancer.
Materials and Methods
Subjects and initial bioinformatics data of Helicobacter pylori
DNA blood samples and gastric biopsies were taken from 10 patients with an age average of ~40 years, from the Andean zone of Tuquerres, and 9 samples from patients from the Pacific coast from the city of Tumaco, Colombia. The sequences were annotated using the prokaryote genome annotation from NCBI. The genomes correspond to sequenced data by our group in collaboration with Valderlbilt University and available on NCBI data base (Table 1) [25-30].
| Risk |
Isolate ID |
Access code |
Diagnosis |
Genome size |
N̡̉ of contigs |
| HRa |
SV328_2 |
MTWO00000000 |
Dysc |
1,645,479 |
56 |
| HR |
SV340_2 |
MTWP00000000 |
NAGd |
1,633,298 |
53 |
| HR |
SV355_2 |
MTWQ00000000 |
IMe |
1,635,304 |
39 |
| HR |
SV376_1 |
MTWR00000000 |
IM |
1,691,791 |
207 |
| HR |
SV380_1 |
MTWU00000000 |
IM |
1,631,819 |
40 |
| HR |
SV397_2 |
MTWS00000000 |
NAG |
1,668,205 |
47 |
| HR |
SV449_1 |
MTWT00000000 |
IM |
1,654,884 |
41 |
| HR |
PZ5056* |
ASYU00000000 |
NAG |
1,578,164 |
335 |
| HR |
PZ5080* |
ASYV00000000 |
IM |
1,597,127 |
283 |
| HR |
PZ5086* |
ASYW00000000 |
NAG |
1,547,845 |
295 |
| LRb |
PZ5005_3A3 |
MTWJ00000000 |
NAG |
1,672,956 |
51 |
| LR |
PZ5006_3A3 |
MTWK00000000 |
NAG |
1,643,170 |
53 |
| LR |
PZ5009_3A2 |
MSYO00000000 |
NAG |
1,677,035 |
53 |
| LR |
PZ5016_3A3 |
MTWL00000000 |
MAGf |
1,644,424 |
44 |
| LR |
PZ5019_3A3 |
MTWM00000000 |
IM |
1,681,561 |
44 |
| LR |
PZ5033_3A2 |
MTWN00000000 |
IM |
1,656,908 |
60 |
| LR |
PZ5004* |
ASZF00000000 |
NAG |
1,569,902 |
303 |
| LR |
PZ5024* |
ASYS00000000 |
NAG |
1,496,849 |
413 |
| LR |
PZ5026* |
ASYT00000000 |
NAG |
1,604,992 |
253 |
Note: aHigh Risk: Host is resident of Tuquerres where risk for gastric cancer is high; bLow Risk: Host is resident of Tumaco where risk for gastric cancer is low; cDys, dysplasia; dNAG, nonatrophic gastritis; eIM, intestinal metaplasia; fMultifocal atrophic gastritis; *Data samples from previous work by Sheh et al. [29].
Table 1: Complete genome description of H. pylori strains in NCBI from two regions of the department of Nari̮̱o, Colombia.
The blood and gastric biopsies were coding as follow, for Tuquerres samples (SV328_2, SV340_2, SV355_2, SV376_1, SV380_1, SV397_2, SV449_1), for Tumaco (PZ5005_3A3, PZ5006_3A3, PZ5009_3A2, PZ5016_3A3, PZ5019_3A3, PZ5033_3A2) (29).
All participants provided informed consent; the study was approved by the institutional and local hospitals review boards. The bioinformatics analysis were performed during January and February of 2021. The human samples were genotyped using an Immunochip previously reported [25], which identifies around 196 × 103 SNPs in genes involved in immune disorders. The Admixture model of STRUCTURE assuming correlated allele frequencies, (50,000 iterations after a burn-in of 50,000 iterations).
The reference populations used in this study were published previously in Human Genome Diversity Project that content European, Amerindian and African ancestries [26,27]. The number of tentative populations (K) was set from 1 to 3 and 10 runs were executed for each K. The STRUCTURE results (mixing model) showed that the model probability was maximized in k=3 [14]. CLUMPP was used to collate replicate runs and calculate means of individual ancestry [28].
To make the phylogenetic modelling we used African, Asian, European and Native American reference genomes [21]. We included genomes from populations of Managua (Nicaragua) and Mexico City (Mexico) belonging to previous study [23]. The genomes from Colombia were 7 isolated from Bogota (CG22366, CA22327, CA22311, CA22339, CA22312, CM22360, CM22351), 10
from Cundinamarca department (CC22402, CC26084, CC26093, CM22346, CA22337, CM22341, CG22389, CG22322, CA22393, CM22388), 14 from Boyaca department (CG22025, CG22087, CG22023, CM22046, CM22013, CG22367, CM22021, CG22370, CM22331, CM22368, CA22020, CM22315, CA22335, CM22347), one from Caldas department (CM22390), One from Caqueta department (CC26100), one from Meta (CA26024), four from Santander department (CG22385, CG22378, CA22019, CA22095) and two from Tolima department (CA22362, CA24004) [22].
Multilocus Sequence Typing (MLST) analysis based on genomes
The housekeeping genes atpA, efp, mutY, ppa, trpC, ureI, and yphC were annotated using PubMLST, and the sequences were selected, downloaded and concatenated. The concatenated sequences were aligned using Muscle software [31]. The phylogenetic analysis was constructed and calculated using a similarity analysis by means of Neighbor-joining [32] with the evolutionary model T92+G+I (Tamura with Gamma variation and invariable sites). The bootstrap analysis was done with 1000 replicates and the phylogenetic tree was edited in iTol v3.
Helicobacter pylori phylogenomic analysis
To the core genome analysis all the sequences were imported from bacteria isolated genome sequences database BIGSdb [33]. Then an alignment of gene by gene was done using H. pylori coding sequences CDS from African strain J99 as reference, and the alignment was exported from the database. The output matrix from the genome comparing obtained by BIGSdb was used to create the phylogenomic tree using MEGA V7 [34].
The phylogenomic analysis based on SNPs was carried out using CSI-phylogeny [35] with the default parameters. The genome assembly was analyzed with the following parameters: minimum depth at SNPs positions of 10; relative depth at SNPs positions of 10; minimum distance between SNPs (prune) of 10; minimum SNPs quality of 30; minimal read mapping quality of 25; minimum Z-score: 1.96 corresponding to a p<0.05 value. The reads were mapped to the reference genome J99 with BMW mem, and the SNPs were assigned with the mpileup tool from SAMTools [36]. The SNPs were filtered according to the assigned parameters to obtain a high-quality matrix. The SNPs matrix was created evaluating all the positions for each genome, which were concatenated creating a multiple FASTA file used in the Maximum- likelihood phylogenetic analysis, where we found 175,856 SNPs. The results were visualized and edited with FigTree v1.4.0 [37].
VacA cytotoxin and AlpA adhesin phylogenetic analysis
A phylogenetic analysis of virulence gene vacA and adhesine gene alpA were studied. The sequences were depurated and aligned using Muscle software [31]. We used the tool Gblocks [38] to determine the parsimony site due to the high diversity of the genes. The evolutive model that better adjusted to the alignment was the General Time Reversible GTR+G+I that shows a Bayesian information criterion BIC=125767.110, lnL=-60531.321. The variation rate between sites was modeled with Gamma distribution=0.57. The analysis involved 196 DNA sequences with long of 2948pb. The vacA gene the phylogenetic analysis was determined, created and calculated using Maximum-likelihood estimation and a bootstrap analysis of 1000 replicates for more statistical accuracy in PhyML v. 3.0 [39].
To the gene alpA the best evolutive model was determined by the General Time Reversible GTR+G+I with BIC=43727.101, lnL=- 19161.291. The variation rate between sites was modeled with a Gamma distribution of 0.66. The analysis involved 215 DNA sequences with long of 1093pb. For the gene alpA we applied the Maximum-likelihood method along with a bootstrap of 1000 replicates in PhyML v. 3.0 [39]. The phylogenetic trees were visualized and edited with FigTree v.1.4.0 [33].
Results
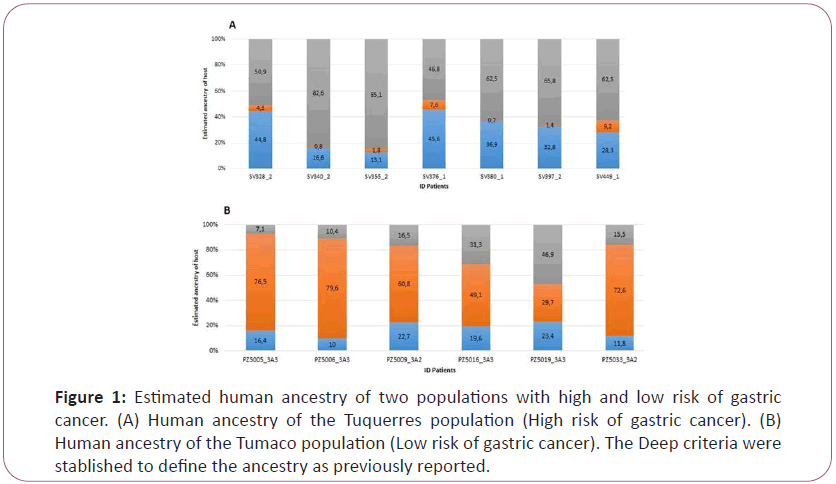
Human ancestry analysis
In Colombia, during European colonization African and Spanish human population arrived then, they met Native American population in the American continent. The results from the human ancestry analysis showed that on in the Andean zone of Tuquerres, the population showed a marked Amerindian and European ancestry (Figure 1A). On the other hand, in the Pacific coast, patients presented a high ethnic combination, a great proportion of African together with an Amerindian and European mixture (Figure 1B), which corroborates the results obtained in a previous work [14].
Figure 1: Estimated human ancestry of two populations with high and low risk of gastric
cancer. (A) Human ancestry of the Tuquerres population (High risk of gastric cancer). (B)
Human ancestry of the Tumaco population (Low risk of gastric cancer). The Deep criteria were
stablished to define the ancestry as previously reported.
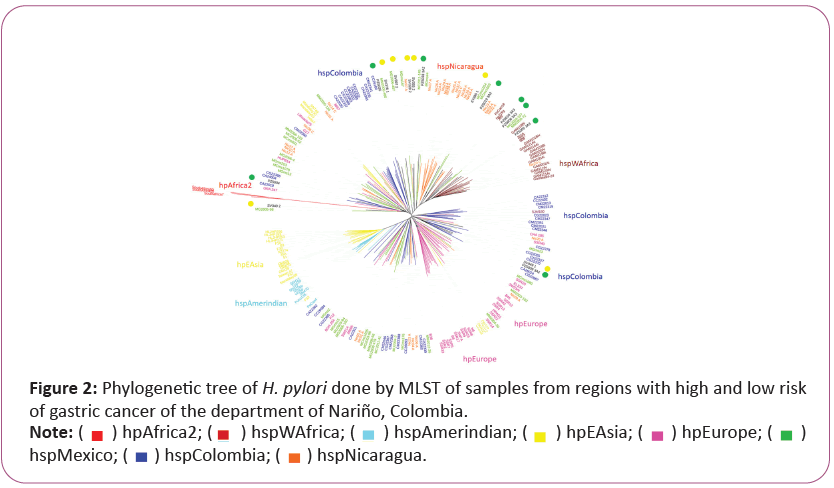
Multilocus Sequence Typing (MLST) analysis based on Helicobacter pylori genomes from Narino, Colombia
In the MLST analysis we observed independent clades of hspWAfrica, hpAfrica2, hpEAsia, hpEurope, hspAmerindian and independent linages of Latin American strains (hspColombia and hspNicaragua). In the clade of Native American we observed a close evolutive relation with Asian strains. In the hpEurope linage, we found isolates from Asia continent that has been reported as ancestors in this populations [15,40]. Likewise, a group of bacteria from Colombia, Mexico and Nicaragua located in the Amerindian and European clades were also mixed with European isolates.
The strains from both study sites (SV449_1 and PZ5009_3A2) showed in the phylogenetic tree association with independent Colombian strains (hspColombia). Although, we observed four strains from the pacific coast (PZ5019_3A3, PZ5016_3A3, PZ5006_3A3, PZ5005_3A3) that along with others from Mexico and Nicaragua were associated to West Africa (hspWAfrica), and interestingly the phylogenetic tree we found a Colombian isolated group composed by an isolate from Pacific coast (PZ5004) and an isolated from Andean zone (SV340-2) that showed association with the most ancestral linage of H. pylori (hpAfrica2) from the South African Continent. A group of four isolates from Tuquerres (SV376_1, SV397_2, SV328_2 and SV355_2) and two from Tumaco (PZ5026, PZ5033_3A2) clustered with Latin American ancestors (Figure 2).
The tree was created by mean of Neighbour-joining. The isolates from Túmaco, Colombia (low risk of gastric cancer) were marked with green point, and those from Túquerres, Colombia (high risk of gastric cancer) were marked with yellow point. The IDs corresponding to each H. pylori isolate were discriminated by color, and they can be seen in the outward perimeter of the tree. The different H. pylori populations (hp) and subpopulations (hsp) are described on the right.
Phylogenomic analysis of Helicobacter pylori confirming the positioning of the isolates from Narino in the groups of Colombian (hspColombia) and hspWAfrica isolates
In the tree obtained from the core phylogenomic analysis we also observed the formation of independent clades of hpEurope, hspAmerindian, hpEAsia, hspWAfrica, hspColombia, hspNicaragua and hspMexico, with hpAfrica2 being the farthest group. These results were consistent with those from the MLST analysis, which showed that hspAmerindian is among the group of bacteria from hpEAsia. From Tuquerres site 8/10 (80%) H. pylori isolates (SV449_1, SV355_2, PZ5080, PZ5086, SV328_2, SV376_1,
SV380_1 and SV397_2) formed an independent linage with Colombian isolates (hspColombia) and only 2/10 (20%) isolates (SV380_1, SV397_2) grouped with African ancestors (hspWAfrica). Regarding the H. pylori isolates from Tumaco 5/9 (56%) (PZ5004, PZ5005_3A3, PZ5024, PZ5016_3A3 and PZ5006_3A3) clustered
with African ancestors (hspWestAfrica). However, 4/9 (44%) isolates (PZ5019_3A3, PZ5033_3A2, PZ5009_3A2 and PZ5026)) grouped with Colombian linage (hspColombia) (Figure 3).
Those in yellow point are from the Andean zone, green point for the Pacific zone. Different H. pylori populations (hp) and subpopulations (hsp) are described on the right.
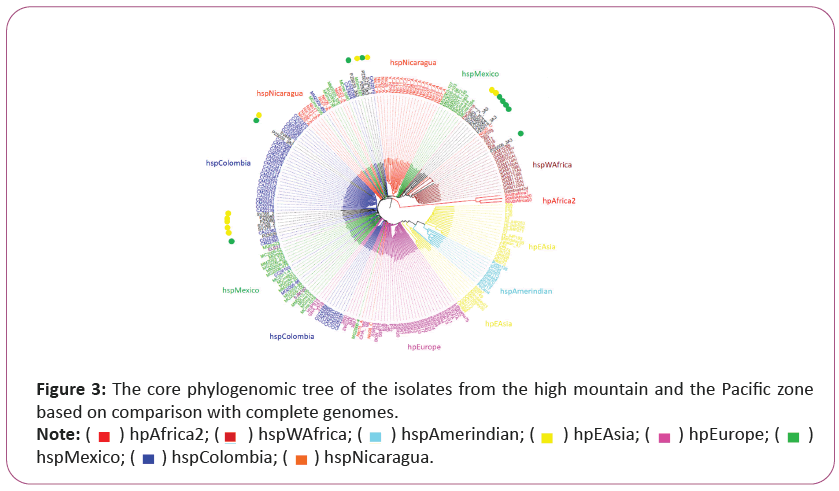
The phylogenomic tree created using SNPs we observed similarly to previous results formation of independent clades for H. pylori from each continent. In addition, we found a group of strains with Colombian and Nicaraguan origin inside the hpEurope. Also, the hpEurope linage showed a close evolutive relation to Asian strains (hpEAsia) as same as the MLST analysis. In the West Africa linage we observed isolated from Mexico, Nicaragua and Colombia. The H. pylori isolates from Tumaco 5/9 (56%) that were reported in the phylogenomic tree based on SNPs genome grouped with hspWAfrica (PZ5006_3A3, PZ5016_3A3, PZ5004, PZ5024, PZ5005_3A3) and 4/9 (44%) H. pylori isolates (PZ5019_3A3, PZ5033_3A2, PZ5009_3A2 y PZ5026) were observed forming an independent linage (hspColombia) (Figure 4).
Those in yellow point are from the Andean zone, green point for the Pacific zone. Different H. pylori populations (hp) and subpopulations (hsp) are described on the right.
From Tuquerres population 8/10 (80%) isolates formed in the phylogenomic tree based on SNPs a cluster with Colombian ancestors (hspColombia) and only 2/10 (20%) isolates (SV397_2 y SV380_1) were grouped with African ancestors (hspWAfrica), (Figure 4). We observed that both used methods showed identical proportions of H. pylori ancestry for both phylogenomic trees in both studied populations.
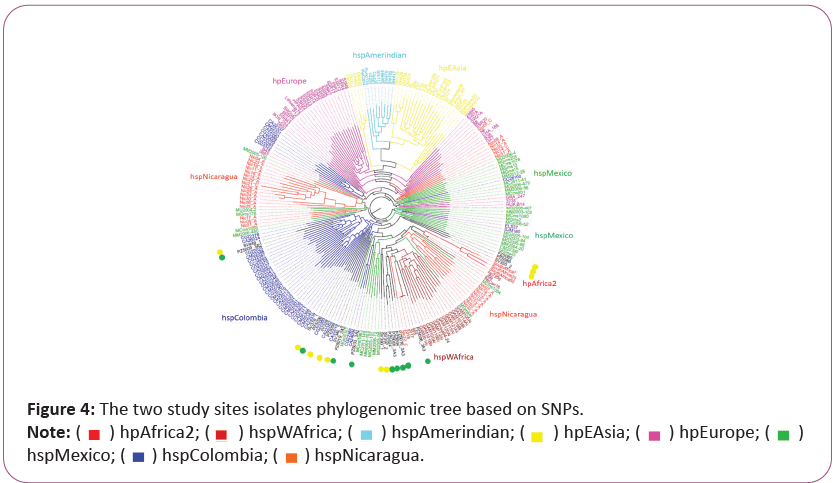
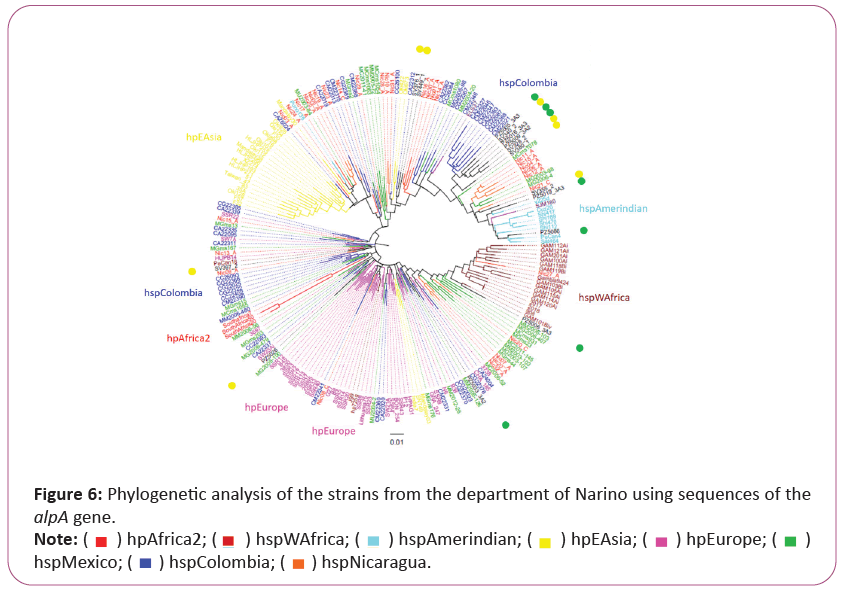
Phylogenetic analysis of vacA and alpA revealing the origin and rapid evolution of Helicobacter pylori isolates from Narino, Colombia
The analyzed nucleotide sequence of the vacA gene showed results where the six isolates from the Andean zone (SV326_2, SV449_1, SV340_2, PZ5080, SV355_2 and PZ5056) and five strains from Pacific coast (PZ5026, PZ5016_3A3, PZ5033_3A2, PZ5019_3A2, PZ5009_3A2) are grouped preferentially with hspColombia. Andean strain SV380_1 indicated a strong relation with Mexican strains. Pacific coast strains PZ5005_3A3 and PZ5006_3A3 were grouped with African strains hspWAfrica. Nevertheless, Amerindian strain PeCan4 was observed with the Mexican strains (Figure 5).
The isolates from Tumaco, Colombia (low risk of gastric cancer) were marked with green point, those from Tuquerres, Colombia (high risk of gastric cancer). Different H. pylori populations (hp) and subpopulations (hsp) are described on the right.
In the alpA gene phylogenetic tree we observed independent linage described previously, it is clear that a group of Colombian and Mexican isolates are very close evolutive related to hpAfrica2 strains. We observed that a group of isolates from both study sites Tumaco (Three strains: PZ5005_3A3, PZ5016_3A3 and PZ5009_3A2) and Tuquerres (Three strains: SV340_2, SV328_2 and SV380_1) were differentiated in the hspColombia clade, although there were strains from the same regions among the hispanoamerican and European strains. Curiously, in this phylogenetic tree we observed the strains SV355_2 from Tuquerres and PZ5019_3A3 and PZ5086 from Tumaco in the group of isolates provenient from Native Americans (hspAmerindian). Only one isolated from Tumaco (PZ5006_3A3) grouped with West African ancestors (hspWAfrica) (Figure 6).
The tree was created by means of Maximum-likelihood. The isolates from Tumaco, Colombia (low risk of gastric cancer) were marked in green point, those from Tuquerres, Colombia (high risk of gastric cancer) were marked in yellow point. Different H. pylori populations (hp) and subpopulations (hsp) are described on the right.
Discussion
Since the first human migration from East Africa approximately 60,000 years ago [5], H. pylori shows in its genome different migration routes and its human host geographic settlements. The first humans in America came from Asia crossing the Bering Strait 12,000 years ago, by the time and the geographic isolation generated a new H. pylori genetic heritage that infected the Native Americans population (hspAmerindian) [15,18,41-44]. However, the recent colonization by Europeans and Africans no more than 500 years ago involved the mixing of a new microorganism’s burden as H. pylori.
The MLST evolutive and phylogenomic analysis of H. pylori showed that the habitants from Andean zone (20%) and Pacific coast (56%) area had some H. pylori strains from West Africa (hspWAfrica) and South Africa continent (hpAfrica2). There is genetic evidence that from those areas came to America African slaves [45]. Curiously, in several isolates from both populations Tuquerres: (80%); Tumaco: (44%) we observed the development of a new linage to Colombian strains (hspColombia). In contrast to our study, there is other study using MLST that reported H. pylori isolates from the same study sites were associated with isolates from European origin (hpEurope) [19].
The results of virulence gene vacA (isolates PZ5005_3A3 and PZ5006_3A3) and adhesine alpA (isolate PZ5006_3A3) from Tumaco indicated ancestral homology with African strains. Interestingly, the alpA gene showed that the strains SV355_2 and PZ5086 from Tuquerres and PZ5019_3A3 from Tumaco were associated with Amerindian origin strains (hspAmerindian). By the other side, we observed that most of the isolates from both study sites were clustered with small groups of hspColombia.
In Narino department, using MLST it is been identified that Native American strains hspAmerind from populations from Andean area were moved by European strains hpEurope in mestizo coming from Amerindian and Spanish ancestors and hspWAfrica in population from Africa, Europe and Amerindian ancestors [14,19,20]. However, the new findings about alpA gene from isolates with ancestral homology allow to understand that there is no had movement or substitution of native strains.
All this suggest multiple colonization in these populations by strains with different phylogeography origin as hpAfrica2, hspWAfrica, hpEurope and hspAmerindian. The multiple origins of those genes could be used as prerequisite to be able of fixation and colonization on gastric mucosa, in response to genetic, immune and physiological host characteristics [21]. In consequence, these genes will be present due to the variability of competitive H. pylori strains that inside a host cooperated through quorum sensing [46,47].
In the Andean region (Tuquerres) the presence of hspColombia cluster could be product of the adaptation to the new ecological niche from mestizo populations. However, the infection by a new subpopulations implies a new encounter between the host and bacteria, in case of a relation no synchronized that favor the genotypes and phenotypes co-evolved, involve the hosts and pathogen extinction [48]. This includes a new set of interactions between virulence factors that provide immune mechanisms evasion systems and colonization by H. pylori. The bacterium performs this two process by two genes, vacA that synthetize a protein that inhibits the host immune response and increase the signaling pathways of gastric mucous cells allowing the insertion of CagA proteins that improve the H. pylori growing [47,49]. The recent gene adaptation probably brought consequences on pathogenesis in Tuquerres, for example, the vacA alleles s1m1 that are associated to major virulence are more frequent in this population and the cagA gene expression is higher in the Andean region than Pacific coast [50,51]. Possibly causing a high H. pylori proliferation and an ecological unbalanced of bacteria communities allowing the pre-cancerous lesion development in human population.
The incidence of gastric cancer rates changes around the world, for example, in several African countries the cancer gastric incidence is low (0,6 per 100,000 inhabitants), even although the H. pylori infection is ubicuos [52-55]. This could be due to that during the hunter-gatherers time in the African savannah, the process of human evolution was like a bottle effect to very pathogenic microorganisms, selecting and transmitting to the next generations only the commensal bacteria. This could extinguish the very pathogenic bacteria with its host, like H. pylori [56]. The arriving of these African populations to Colombian pacific coast brought H. pylori commensal strains that have co-evolved with human host during thousands of years leading to less probability of disease development [14].
It is important to mention that the Th immune response plays an important role in gastric cancer pathogenesis [12]. The response is multifactorial against a specific strain [14]. This has been observed in the African continent where the Th2 response is predominant, while in Japan (high gastric cancer incidence) the immune response is Th1 type. These differences have been related with a minor gastric cancer incidence on Africa [56].
In both study regions the coinfection with other microorganisms like helminths has modulated the immune response [56]. In Tumaco population where the human ancestry is the African origin mainly, it is observed an immune response patron Th2, where the co-infection with helminths is a factor that has modulated this mechanism [57]. The Th2 response is an anti-inflammatory type as immune mechanism to infection by H. pylori strains of African ancestry, product of a co-evolution period since the H. sapiens evolved from a common African branch in parallel to H. pylori since more than 110,000 years [14,19,20].
However, in Tuquerres population the infection by H. pylori of hspColombia linage that causes an immune response Th1 pro- inflammatory [14,19,57], which interaction with human host has happened since about 500 years, short co-evolution and adaptation time after European and African colonization, that suggest evidence of high disease risk in Andean area from Narino department.
In addition, other studies have reported that the dietary differences between the mountain and coast regions influenced the incidence rates, for example, in the coast region there are high intake of fresh vegetables and fruits rich in antioxidants and seafood. While in Andean regions the dietary regimen consists on potatoes and broad beans [12,55,58].
We acknowledge that our study has a low isolates number that limits the correlation statistically the strain origin with gastric lesion type. Also, our isolates number limits us to corroborate with whole genome analysis if the strains with African origin in human population with high Amerindian ancestry produce a higher risk of carcinogenesis as found by MLST in H. pylori and human ancestry as in a previous study [14]. Also, the low number of H. pylori isolates in both study areas limits our study related with the mutation or variants analysis that give major capability of colonization and virulence of H. pylori.
Conclusion
The perspectives to overcome our study limitations are related with the enrollment of new population in Narino with high gastric cancer risk. However, this study gives a preliminary knowledge about H. pylori evolution with mixed human host from Colombia. Also, this study could be a good example to understand the mixing process in other bacteria after the European and African colonization in America territory.
Acknowledgements
We would like to thank the reviewers for the constructive comments and guidelines, and also the Universidad de Narino for the availability of the server for the evolutionary analyses.
Funding Source
This work was funded by the Grupo de Salud Publica and the Vicerrectoría de Investigaciones e
Interacción Social from the Universidad de Narino.
Competing Interests
The authors declare that they have no competing interests.
References
- Peek RM, Blaser MJ (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2:28-37.
- Cover TL, Blaser MJ (2009) Helicobacter pylori in health and disease. Gastroenterology 136:1863-1873.
- International Agency for Research on Cancer (1994) Schistosomes, liver flukes and Helicobacter pylori. IARC working group on the evaluation of carcinogenic Risks to Humans. Risks Hum 61:1–279.
- Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, et al. (2012) Age of the association between Helicobacter pylori and man. PLoS pathogens 8:1002693.
- Linz B, Balloux F, Moodley Y, Manica A, Liu H, et al. (2007) An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915-918.
- Atherton JC, Blaser MJ (2009) Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Investig 119:2475-2487.
- Jones KR, Whitmire JM, Merrell DS (2010) A tale of two toxins: Helicobacter pylori CagA and VacA modulate host pathways that impact disease. Front Microbiol 1:115.
- Gangwer KA, Shaffer CL, Suerbaum S, Lacy DB, Cover TL, et al. (2010) Molecular evolution of the Helicobacter pylori vacuolating toxin gene vacA. J Bacteriol Res 192:6126-6135.
- Odenbreit S, Till M, Hofreuter D, Faller G, Haas R (1999) Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol 31:1537-1548.
- Lu H, Wu JY, Beswick EJ, Ohno T, Odenbreit S, et al. (2007) Functional and intracellular signaling differences associated with the Helicobacter pylori AlpAB adhesin from Western and East Asian strains. J Biol Chem 282:6242-6254.
- Torres J, Correa P, Ferreccio C, Hernandez-Suarez G, Herrero R, et al. (2013) Gastric cancer incidence and mortality is associated with altitude in the mountainous regions of Pacific Latin America. Cancer Causes Control 24:249-256.
- Correa P, Piazuelo MB (2010) Gastric cancer: the Colombian enigma. Revista Colombiana de Gastroenterologia 25:334-337.
- Matta AJ, Pazos AJ, Bustamante-Rengifo JA, Bravo LE (2017) Genomic variability of Helicobacter pylori isolates of gastric regions from two Colombian populations. World J Gastroenterol 23:800.
- Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, et al. (2014) Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci 111:1455-1460.
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, et al. (2003) Traces of human migrations in Helicobacter pylori populations. Science 299:1582-1585.
- Devi SM, Ahmed I, Francalacci P, Hussain MA, Akhter Y, et al. (2007) Ancestral European roots of Helicobacter pylori in India. BMC Genom 8:1-1.
- Moodley Y, Linz B, Yamaoka Y, Windsor HM, Breurec S, et al. (2009) The peopling of the Pacific from a bacterial perspective. Science 323:527-530.
- Yamaoka Y (2009) Helicobacter pylori typing as a tool for tracking human migration. Clin Microbiol Infect 15:829-834.
- de Sablet T, Piazuelo MB, Shaffer CL, Schneider BG, Asim M, et al. (2011) Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut 60:1189-1195.
- Shiota S, Suzuki R, Matsuo Y, Miftahussurur M, Tran TT, et al. (2014) Helicobacter pylori from gastric cancer and duodenal ulcer show same phylogeographic origin in the Andean region in Colombia. PloS one 9:105392.
- Gutiérrez-Escobar AJ, Trujillo E, Acevedo O, Bravo MM (2017) Phylogenomics of Colombian Helicobacter pylori isolates. Gut Pathog 9:1-9.
- Muñoz-Ramírez ZY, Mendez-Tenorio A, Kato I, Bravo MM, Rizzato C, et al. (2017) Whole genome sequence and phylogenetic analysis show Helicobacter pylori strains from Latin America have followed a unique evolution pathway. Front Cell Infect Microbiol 7:50.
- Thorell K, Yahara K, Berthenet E, Lawson DJ, Mikhail J, et al. (2017) Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas. PLoS Genet 13:1006546.
- Correa P, Piazuelo MB (2012) Evolutionary history of the Helicobacter pylori genome: implications for gastric carcinogenesis. Gut and liver 6:21-28.
- Cortes A, Brown MA (2011) Promise and pitfalls of the Immunochip. Arthritis Res Ther 13:1-3.
- International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320.
- The 1000 Genomes Project Consortium (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65.
- Jakobsson M, Rosenberg NA (2007) CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801-1806.
- Pazos A, Kodaman N, Piazuelo MB, Romero-Gallo J, Sobota RS, et al. (2017) Draft genome sequences of 13 Colombian Helicobacter pylori strains isolated from Pacific Coast and Andean residents. Genome Announc 5: 113-117.
- Sheh A, Piazuelo MB, Wilson KT, Correa P, Fox JG (2013) Draft genome sequences of Helicobacter pylori strains isolated from regions of low and high gastric cancer risk in Colombia. Genome Announc 1.
- Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792-1797.
- Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425.
- Rambaut A (2009) FigTree. Tree figure drawing tool.
- Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinform 25: 1451-1452.
- Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147-164.
- Jolley KA, Maiden MC (2010) BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC bioinformatics 11: 1-1.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger data sets. Mol Biol Evol 33: 1870-1874.
- Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O (2014) Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PloS One 9: 104984.
- Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987-2993.
- Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564-577.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59: 307-321.
- Maixner F, Krause-Kyora B, Turaev D, Herbig A, Hoopmann MR, et al. (2016) The 5300-year-old Helicobacter pylori genome of the Iceman. Science 351: 162-165.
- Kersulyte D, Kalia A, Gilman RH, Mendez M, Herrera P, et al. Helicobacter pylori from Peruvian amerindians: traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PloS one 5: 15076.
- Kersulyte D, Bertoli MT, Tamma S, Keelan M, Munday R, et al. Complete genome sequences of two Helicobacter pylori strains from a Canadian arctic Aboriginal community. Genome Announc 3: 209-215.
- Salas A, Richards M, Lareu MV, Scozzari R, Coppa A, et al. (2004) The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet 74(3): 454-465.
- Lu H, Hsu PI, Graham DY, Yamaoka Y (2005) Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology 128: 833-848.
- Argent RH, Hale JL, El-Omar EM, Atherton JC (2008) Differences in Helicobacter pylori CagA tyrosine phosphorylation motif patterns between western and East Asian strains, and influences on interleukin-8 secretion. J Med Microbiol 57: 1062-1067.
- Linz B, Windsor HM, Gajewski JP, Hake CM, Drautz DI, et al. (2013) Helicobacter pylori genomic microevolution during naturally occurring transmission between adults. PloS One 8: 82187.
- Aras RA, Kang J, Tschumi AI, Harasaki Y, Blaser MJ (2003) Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc Natl Acad Sci 100: 13579-13584.
- Bravo LE, van Doorn LJ, Realpe JL, Correa P (2002) Virulence-associated genotypes of Helicobacter pylori: do they explain the African enigma? Am J Gastroenterol Suppl 97: 2839-2842.
- Loh JT, Shaffer CL, Piazuelo MB, Bravo LE, McClain MS, et al. (2011) Analysis of cagA in Helicobacter pylori strains from Colombian populations with contrasting gastric cancer risk reveals a biomarker for disease severity. Cancer Epidemiol Biomark Prev 20: 2237-2249.
- Mitchell HM, Ally R, Wadee A, Wiseman M, Segal I (2002) Major differences in the IgG subclass response to Helicobacter pylori in the first and third worlds. Scand J Gastroenterol 37: 517-522.
- Holcombe C (1992) Helicobacter pylori: the African enigma. Gut 33: 429-431.
- Campbell DI, Warren BF, Thomas JE, Figura N, Telford JL, et al. (2001) The African enigma: low prevalence of gastric atrophy, high prevalence of chronic inflammation in West African adults and children. Helicobacter 6: 263-267.
- Piazuelo MB, Correa P (2013) Gastric cancer: Overview. Colomb Med 44: 192-201.
- Blaser MJ, Kirschner D (2007) The equilibria that allow bacterial persistence in human hosts. Nature 449: 843-849.
- Piazuelo MB, Camargo MC, Mera RM, Delgado AG, Peek Jr RM, et al. (2008) Eosinophils and mast cells in chronic gastritis: Possible implications in carcinogenesis. Hum Pathol 39: 1360-1369.
- Camargo MC, Burk RF, Bravo LE, Piazuelo MB, Hill KE, et al. (2008) Plasma selenium measurements in subjects from areas with contrasting gastric cancer risks in Colombia. Arch. Med. Res 39: 443-451.

 hpEurope;
hpEurope; hspMexico;
hspMexico;