Review Article - (2023) Volume 7, Issue 3
The Overview of CRISPR Technology and its Application on Farm Animals
Vinay Kumar Mehra* and
Satish Kumar
Department of Animal Biotechnology, National Dairy Research Institute, Tehran, Iran
*Correspondence:
Vinay Kumar Mehra, Department of Animal Biotechnology, National Dairy Research Institute, Tehran,
Iran,
Email:
Received: 06-Apr-2020, Manuscript No. IPJASLP-23-4516;
Editor assigned: 10-Apr-2020, Pre QC No. IPJASLP-23-4516(PQ);
Reviewed: 24-Apr-2020, QC No. IPJASLP-23-4516;
Revised: 02-May-2023, Manuscript No. IPJASLP-23-4516(R);
Published:
30-May-2023, DOI: 10.36648/2577-0594-7.2.41
Abstract
Livestock animals are important for agriculture economy and biomedical research. They are sources of mike, meat, egg, leather and other products. The development of genome editing technologies, especially CRISPR-Cas have revolutionized the generation of gene edited farm animals. In this review, we briefly introduce the CRISPR-Cas technology and highlight its application on livestock such as human disease modeling, disease resistant animal, alteration of milk composition, animal welfare and other agricultural and biomedical related traits which enhance the livestock production in order to meet the increasing demand of food worldwide. Besides the several benefits of CRISPR-Cas technology, the risk factors and ethics issues related to this technology should be reconsidered before enter into the CRISPR era.
Keywords
CRISPR; Genome editing; Livestock; Disease model; Cow milk allergy
Introduction
Large animals play important roles in biomedical research.
Animal models are crucial for understanding disease
pathogenesis and developing novel therapeutic drug and
treatments. Livestock animals are important sources of milk,
meat, leather and other products. The recently developed
genome editing techniques have been used for generation of
gene-modified large animals that are used for biological,
biomedical and agricultural research and increase the
performance of the animals in terms of both qualitative and
quantitative. The gene-edited livestock animal has been used
as a bioreactor for the production of human biological
products such as transgenic pig with human albumin in their
blood [1].
Genome engineering is defined as direct manipulation into
the genome that can make specific edition/deletion to the genome by engineered nucleases, which offer a perfect
platform to knock out/in and replace the particular DNA
fragment, and make accurate genome editing on the genome
level. There are three major types of programmed nucleases:
Zinc Finger Nucleases (ZFNs), Transcription Activator Like
Effector Nucleases (TALENs) and the Clustered Regularly
Interspaced Short Palindromic Repeat-associated nuclease
Cas9 (CRISPR-Cas9). These nucleases produced DNA Double-
Strand Breaks (DSBs) at specific sites in the genome by
targeted recognition and cleavage. Out of these CRISPR/Cas9
system is the latest genome editing technology, which are
most commonly used nowadays. CRISPR is part of the
bacterial genome system, which makes the bacterial cells
immune to virus [2]. The CRISPR was first observed in Escherichia coli and served as an adaptive immune system in
bacteria against bacteriophages. CRISPR systems, found in
90% of archaeal and 40% of bacterial genomes are highly diverse, with variation in Protospacer Adjacent Motif (PAM)
sequences and the number and type of Cas proteins. The
CRISPR system has three types of mechanisms i.e type I, II and
III. In type I and type III CRISPR, various types of Cas proteins
are participated in the recognition and destruction of target.
However, in Type II CRISPR/Cas9 system low number of Cas
proteins are involved, so thereby engineering of type II CRISPR
system much simpler [3].
The main question is that why genome editing technology
should be used in livestock. Although conventional methods,
such as management of animal health, nutrition and
reproduction, make an important contribution to improving
the productivity of the animals. However, increasing the
demand of food allover growing world, these approaches are
not enough to meet the demands, so CRISPR/Cas9 technology
emerges as a powerful tool to overcome this problem
globally. So the main aim of this review paper is to describe
the CRISPR technology and its application in livestock animals
[4].
Literature Review
The CRISPR/Cas9 System
The CRISPR/Cas9 system is a part of adaptive immune system
of the bacteria. For the first time showed that this system
could be designed as genome editing tool in vitro and in 2013
the CRISPR/cas9 genome editing tool demonstrated on the
human and mice cells [5]. The CRISPR/Cas9 system contains
three main components: Endonuclease (Cas9), CRISPR RNA
(crRNA) and trans-activating crRNA (tracrRNA). The Cas9
endonuclease which belongs to Type II CRISPR/Cas9 system,
cut the target sites with the help of two RNAs: The crRNA
which assign the genomic target for Cas9 and the tracrRNA
which serve as a scaffold linking the crRNA to Cas9 and help in
processing of maturation of crRNAs into a small single RNA
sequence known as the guide RNA (gRNA) or single guide RNA
(sgRNA). The specific recognition of target site by sgRNA
depends on the Protospacer Adjacentmotifs (PAM), which is
located downstream of 3′ end of the target sequence and
after recognition of target site by guide RNA, the Cas9
produces a Double Stand Break (DSB) on the complementary
locus [6]. The generated DSBs will be repaired by two
pathways, Non-Homologous End Joining (NHEJ) and
Homology-Directed Repair (HDR) (Figure 1).
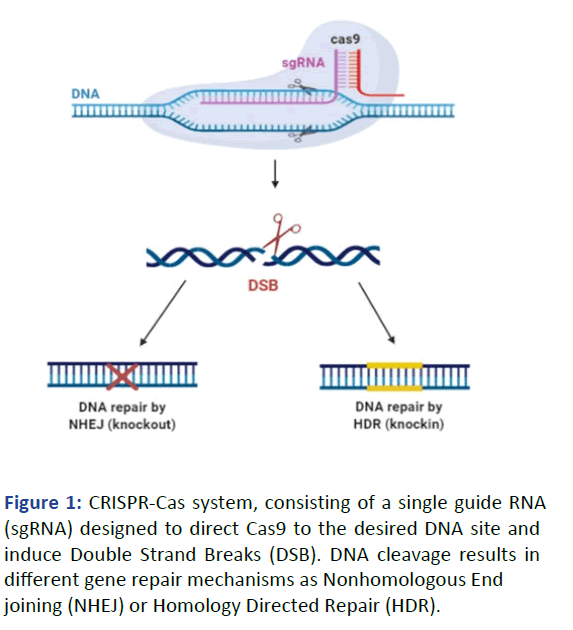
Figure 1: CRISPR-Cas system, consisting of a single guide RNA
(sgRNA) designed to direct Cas9 to the desired DNA site and
induce Double Strand Breaks (DSB). DNA cleavage results in
different gene repair mechanisms as Nonhomologous End
joining (NHEJ) or Homology Directed Repair (HDR).
The NHEJ mechanism caused small indels or chromosomal
rearrangements and disrupts open reading frames, which
leads to knockout of the gene. In case of HDR pathway DSB is
repaired by the through sequence homology. Compare to
HDR, the NHEJ is more active since HDR mechanism needs a
homologous template and is mainly restricted to S and G2
phases of the cell cycle [7,8].
The Effec ive Endonuclease
For any CRISPR experiment one of the main things is to
determine the suitable endonuclease [9]. The spCas9
(Streptococcus pyogenes) nuclease is most commonly used
but several other type of nucleases such as Cas12a or CasX
are also present ,which have some unique properties and can
be used depending on the objective of the project. There are
two domains in the Cas9 nuclease: HNH and RuvC-like
domain. The HNH nuclease domain cut the target strand of
the nucleotide and RuvC-like domain cut the noncomplementary
strand. To increase the specificity and
adaptability of the Cas9, researcher reprogrammed the Cas9
into dCas9 nuclease (dead Cas9). The dCas9 do not have the
DNA cleaves activity but can still bind to the target site with
the help of guide RNA (gRNA) [10]. However the CRISPR/
dCas9 system cannot insert or delete the gene, instead it
silences the target gene by blocking the transcription process,
while CRISPR/Cas9 system produced DSB on the target site. So
the CRISPR/dCas9 tool can be used for genetic screening,
gene regulation and epigenetic regulation. Additionally, the
newly RNP based CRISPR tools have also been developed, in
which different Cas RNA with sgRNA used for genome editing
[11].
The single guide RNA (sgRNA)
The second important component of CRISPR/Cas9 system to
edit the genome is selection and design of sgRNA [12].
Generally three rules considered during designing of sgRNA;
the sgRNA should have
• Fewest potential off-target matches.
• Highest predicted cleavage efficiency.
• Target early exons these rules increase the probability of
frameshift and nonsense mutations that disrupt gene
expression.
The target sites for sgRNA are generally 18 bp-20 bp long and
designed immediately to upstream of a Protospacer Adjacent
Motif (PAM). The PAM sequence varies depending upon the
type of cas nuclease. The PAM for spCas9 is 5’-NGG-3’ and the
PAM for saCas9 is 5’-NNGRRT-3. There are many online
CRISPR tools and resources are available to design the sgRNA,
some of this are list in Table 1.
| Name |
URL |
| Addgene |
http://www.addgene.org/crispr/ |
| Benchling |
https://benchling.com/crispr |
| Breakingcas |
http://bioinfogp.cnb.csic.es/tools/breakingcas/ |
| Broad institute GPP |
https://portals.broadinstitute.org/gpp/public/ |
| Chopchop |
http://chopchop.cbu.uib.no/ |
| Crispor |
http://crispor.net/ |
| Crisflash |
https://github.com/crisflash/crisflash |
| Deskgen |
https://www.deskgen.com |
| E-crisp |
http://www.e-crisp.org/E-CRISP/ |
| Microsoft research crispr |
https://crispr.ml |
| RGEN tools |
http://www.rgenome.net/ |
| Synthego |
http://design.synthego.com |
| WTSI genome editing |
https://www.sanger.ac.uk/htgt/wge/ |
Table 1: List of CRISPR guide RNA designing tool.
These softwares analyze the target sequence and identify all
possible 18 bp-20 bp sequences which are immediately after
the PAM sequence (5’-NGG) and provide the essential
information require for selection of sgRNAs. In livestock, the
gestation length is long, so after selecting the desired gRNA
for target sequence, it is highly recommended to confirm
them before producing the edited animals [13]. This analysis
can be done in in vitro using cultured cells of the same species
or directly in embryos.
Delivery of CRISPR System
There are mainly three approaches to deliver the CRISPR
system in vitro or in vivo
• Plasmid encoding both the Cas9 protein and the guide RNA.
• Cas9 mRNA with guide RNA.
• Cas9 protein with guide RNA.
The vehicles used for delivery of CRISPR/Cas9 system are
divided into three groups: Physical delivery, viral vectors and
non-viral vectors. The most common physical delivery
methods are microinjection and electroporation.
In case of viral vectors delivery method, specifically
engineered Adeno-Associated Virus (AAV), and lenti-virus are
used as vehicles. The microinjection delivery method has
highest efficiency compare to other CRISPR/Cas9 delivery
methods [14]. The mail problem for generation of edited
livestock animal is the in vivo zygote production. In vitro Embryo Production (IVEP) is the preferable method for
microinjection delivery of the CRISPR component. In
microinjection delivery method CRISPR system is directly
injected into the cytoplasm of the zygote, thus this method is
fast and more efficient and preferable method in large
animals. The second most preferable method for CRISPR
system delivery in livestock is the SCNT (somatic cell nuclear
transfer) approach; however the efficiency is low compare to
microinjection [15].
CRISPR applications on livestock: Most of the biological and
biomedical research is carrying out on rodents but validation
and preclinical assessment are performed on large animals. In
the last few years, the nuclease-mediated gene editing
technologies has been used in livestock breeding; however
these approaches are more complicated. The development of
CRISPR technology makes the genome editing in large animal easier. Various livestock animal have been produced by using
CRISPR technology during the last few years [16]. The major
applications of CRISPR technology in farm animals are to
increase the productivity like milk, meat and egg, produce
disease resistant animal, production of therapeutic proteins
into the milk and production of human organs for
transplantation which can be describes as follows (Figure 2).
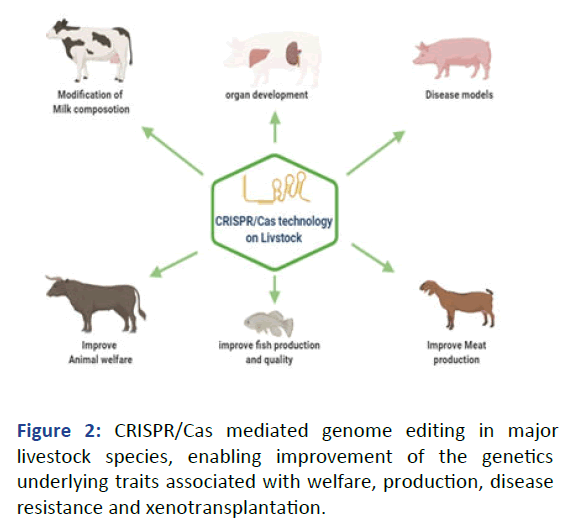
Figure 2: CRISPR/Cas mediated genome editing in major
livestock species, enabling improvement of the genetics
underlying traits associated with welfare, production, disease
resistance and xenotransplantation.
Modificaion of Milk Composition
β-Lactoglobulin (BLG) is a major whey protein of ruminant
milk and is considered as allergens when it is devoid of iron in
the milk and responsible for cow milk allergy disease. Several
methods have been used to diminish the allergenicity of the
BLG, including heat treatment, fermentation, hydrolyzed
protein desensitization, glycation, however these methods
are not so effective. A possible option have been attempted
to disrupt the β-lactoglobulin gene expression by targeting
BLG gene. In 2014 Ni have successfully edited the β-
lactoglobulin gene by using CRISPR/Cas9 system in goats [17, 18]. Similarly also knock out the β-lactoglobulin gene in goat
through CRISPR/Cas9 technique and reduced the expression
of β-lactoglobulin in milk. The BLG knock out cattle has also
been produce by using Zinc-Fnger Nuclease (ZFNs) mRNA but
till now no CRISPR edited BLG knock out cattle has been
produced. In a study human Lactoferrin (hLF) gene was
knocked in at BLG locus through TALEN approach in goat.
These edited goats produce the human lactoferrin, a
glycoprotein involved in iron adsorption and in non-specific
immune reactions in the intestinal tract (Table 2).
| Species |
Gene |
Modifications |
Applications |
| Pig |
Npc1l1 |
KO |
Disease model for cardiovascular and metabolic diseases |
| Pig |
ApoE/LDLR |
KO |
Disease model for cardiovascular diseases |
| Pig |
vWF |
KO |
Disease model for vWD |
| Pig |
TP53/PTEN/APC |
KO |
Disease model for lung cancer |
| Pig |
TPH2 |
KO |
Disease model for 5-HT deficiency induced behavior abnormality |
| Pig |
Huntingtin |
KI |
Disease model for HD |
| Pig |
ApoE |
KO |
Disease model for cardiovascular disease |
| Dog |
MSTN |
KO |
Improve muscle growth, new strains |
| Dog |
GGTA1/CMAH |
KO |
Xenotransplantation |
| Pig |
PERV |
KO |
Xenotransplantation |
| Goat |
MSTN |
KO |
Goat meat production, composition and quality |
| Goat |
MSTN (fat-1) |
KI |
Goat meat production,composition and quality |
| Pig |
UCP1 (mouse UCP1) |
KI |
Pig meat production, composition and quality |
| Pig |
CD163 |
KO |
Disease resistance to PRRSV |
| Cattle |
NRAMP1 |
KO |
Disease resistance to tuberculosis |
Note: KO-Knock Out, KI-Knock In
Table 2: List of CRISPR/Cas9 edited livestock animals for the different purposes.
Discussion
Improvement of Meat Production and Reproduction
Performance
The well-known example related to meat production is
knockout of the Myostatin gene (MSTN). This gene secreted a
myostatin protein in muscle tissues, which act as a negatively
regulator of muscle growth [19]. The mutation in MSTN gene
leads to double-muscling phenotype in the animals and this
was, first reported in cattle and then in sheep, dogs and
humans. The cattle Belgian blue and piedmontese or Texel
sheep are the well-known example of natural mutation in
NSTN gene. So making double-muscling phenotype is an
attractive target for genome editing to increase the meat
production in livestock. In MSTN-mutant Meishan pigs
through Zinc Finger Nucleases (ZFN) technology that showed
an increase in muscle mass by 100% and a decrease in fat
deposition compared to wild-type. In 2018 A CRISPR edited
MSTN knock out goad has been produce, which showed
significantly higher weight gain than that of Wild-Type (WT)
goats. In the same year Zhang et al., knock out the MSTN
gene and then knock-in the fat-1 gene into the goat MSTN
locus by using CRISPR technology. The fat-1 gene product
converts n-6 PUFA (n-6 Polyunsaturated Fatty Acid) into n-3
PUFA, so this CRISPR edited goat showed improved muscle
growth and also produced nutritious meat by decreasing the
ratio of n-6 PUFA to n-3 PUFA, which has been reported as a
risk factor for many human diseases.
The CRISPR technology has also been used for improvement
of thermoregulation in livestock. Uncoupling Protein 1 (UCP1)
play an important role in thermoregulation. The functional
UCP1 protein is absent in pig, which makes them liable to
cold and prone to fat deposition and results in increase the
mortality rate neonates and decreased production efficiency.
Knock-in the UCP1 gene in pig through CRISPR technology
which showed an improved ability to maintain body
temperature during acute cold exposure and also showed
reduced fat deposition, which make them valuable resource
for the pig industry that can improve pig welfare and reduce
economic losses. In 2017, knock out the NANOS2 gene in
domestic pigs by using CRISPR/Cas9 technology to generate
offspring with monoallelic and biallelic mutations. They found
that NANOS2 knockout boar lack a germ line phenotype but
other form of testicular development were normal. These
NANOS2-null boars may provide a good environment to host
germ cells from a genetically superior male, and thus broaden
his genetic potential.
Mycobacterium bovis which caused tuberculosis in livestock
animal especially in cattle is becoming a major risk to the
agricultural and public health which causes significant
economic loss to the farmers. The NRAMP-1 (Natural
Resistance-Associated Macrophage Protein-1) gene provided
innate resistance to intracellular pathogens such as Mycobacterium, Leishmania, Salmonella and Brucella. This
NRAMP-1 gene has been inserted into the cow genome
through CRISPR technology and makes the cow resistant to
tuberculosis. Porcine Reproductive and Respiratory Syndrome
(PRRS) is the most economically important disease of swine all
over the world, which is cause by Porcine Reproductive and
Respiratory Syndrome Virus (PRRSV). The vaccines have been
developed for this disease but unable to control the disease.
The CD163 is the receptor for entry of PRRSV into cells. So in
pig the CD163 protein encoding gene has been knock out
through CRISPR technique, results in no clinical signs of PRRS
were observed in edited pigs which demonstrated that a
single gene deletion creates PRRSV resistant pigs. Niemann-
PickC1-Like-1 (NPC1L1) protein is potent cholesterol
absorption inhibitor that lowers blood cholesterol in humans
and highly expressed in human liver. The NPC1L1 gene has
been introduced into the pig genome to produce animal
mode for understanding of cardiovascular and metabolic
diseases.
Biomedical Application
The most crucial issue for the patients awaiting organ for
transplantation is the availability and rejection of the
allograft. Xenotransplantation is one of the solution in which
the transplantation of animal cells, tissues or organs could
replace an injured tissue or whole organ in humans. Pigs are
the best choice for human organ development because
anatomical and physiological similarities to the human organs
and are cost-effective in breeding but the major problems are
the immune rejection and potential cross species infection. So
to overcome these problems scientists have been introduce
human organ development gene into the pig genome to
produce human organs into the pig. Wu successfully
inactivated the pancreatogenesis in pig embryos via zygotic
co-delivery of Cas9 mRNA and dual sgRNAs targeting the
PDX1 gene, which makes the pigs suitable for the xenogeneration
of human tissues and organs.
Generating B cell deficient mutant is the first step to produce
human antibody repertoires in large animal models. IgM
heavy chain gene is crucial for B cell development and differentiation. This IgM heavy chain gene has been knock-out
in pig through CRISPR technology, resulted in a B cell-deficient
pig. This study highlights the potential of CRISPR technology in
pig, which can used as animal model for study of human
diseases. Human Serum Albumin (HSA) is the most abundant
plasma protein that plays important role in homeostatic
functions in human physiolog. Several approaches have been
used to produce recombinant Human Serum Albumin (rHSA),
however not so effective because of separation and
purification of the rHSA are problematic. To overcome these
problems Peng knocking human albumin cDNA into swine
albumin locus to produce rHSA in pigs, which can used as
bioreactor for the production of human albumin.
The Neuronal Ceroid Lipofuscinoses (NCLs/CLN1) are a group
of inherited, neurodegenerative, lysosomal storage disorders
that affect children and young adults. The more critical
infantile forms of this disease are due to mutations in the
Palmitoylprotein Thioesterase 1 (PPT1) gene, which reduces
the child’s lifespan to approximately 9 years of age. Eaton
produced disease-causing PPT1 (R151X) human mutation into
the orthologous sheep locus through CRISPR/Cas9 technology
and generated a sheep model for the study of CLN1 disease.
Application in aquaculture: The fish farming contributes
billions of dollars to the world economy. The genome editing
technology has also been demonstrated in area. Escaping of
farmed salmon fish into wild stocks is a major threat to the
genetic integrity of wild populations. To overcome this
problem dnd-gene (required for germ cell survival in
vertebrates) has been knock-out by using CRISPR-Cas9 system.
Similarly genome edited ridgetail white prawn has been
produced by microinjection of Cas9 mRNA.
Application in animal welfare: The horned animal in the farm
caused many problem such as they injure the animal, require
more space for feeding, are dangerous to handle and
transport than hornless animals. To over these problem
farmers generally destruct the horn-producing cells before
they grow and attach to the skull (disbudding) to prevent horn
growth. But this practice is very painful to the animal. In 2016
Carlson et al., has been produced the hornless dairy cattle by
Transcription Activator-Like Effector Nucleases (TALENs)
approach but no such type of animal has been produce
through CRISPR technology which is more efficient and easy
technique compare to other genome editing technology.
Conclusion
Genome editing by CRISPR/Cas9 system has become a
prevailing tool in biological research. It have the ability to
generate site-specific indels to the genome, which allows help
in the identification of gene function, allelic variants between
breeds or species, or to create novel phenotypes. Compare to
other genome editing techniques (ZFN and TALEN), CRISPR/
Cas9 system is more efficient and easy to handle. The CRISPR
technology has been successfully used for generation of many
valuable animals for human disease models, xenotransplantation
and the agricultural economy such as
improves the milk composition, meat production and disease resistant livestock animals. Mostly this tool used on the pig
because of anatomical and physiological similarities to
human, however very less effort has been made on cattle.
Buffalo contribute more than 50% milk production in India
and like cow milk; buffalo milk also contains betalactoglobulin
which act as allergen in many children and
adults. So to overcome this problem, CRISPR technology could
be used for the production of beta-lactoglobulin knock out
buffalo in the future.
Conflict of Interest
The authors declare no conflict of interest.
Data Availability
The data that support the findings of this study are available
from the corresponding author upon reasonable request.
References
- Adli M (2018) The CRISPR tool kit for genome editing and beyond. Nat Commun. 9(1):1911.
[Crossref] [Google Scholar] [PubMed]
- Petersen B (2017) Basics of genome editing technology and its application in livestock species. Reprod Domest Anim. 52:4-13.
[Crossref] [Google Scholar] [PubMed]
- Carlson DF, Lancto CA, Zang B, Kim ES, Walton M, et al. (2016) Production of hornless dairy cattle from genome-edited cell lines. Nat Biotechnol. 34(5):479-481.
[Crossref] [Google Scholar] [PubMed]
- Chen F, Wang Y, Yuan Y, Zhang W, Ren Z, et al. (2015) Generation of B cell-deficient pigs by highly efficient CRISPR/Cas9-mediated gene targeting. J Genet Genomics. 42(8):437-444.
[Crossref] [Google Scholar] [PubMed]
- Cho SW, Kim S, Kim JM, Kim JS (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature Biotech. 31(3):230-232.
[Crossref] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science. 339(6121):819-823.
[Crossref] [Google Scholar] [PubMed]
- Cui C, Song Y, Liu J, Ge H, Li Q, Huang H (2015) Gene targeting by TALEN-induced homologous recombination in goats directs production of β-lactoglobulin-free, high-human lactoferrin milk. Sci Rep. 5(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Davis PJ, Smales CM, James DC (2001) How can thermal processing modify the antigenicity of proteins? Allergy. 56:56-60.
[Crossref] [Google Scholar] [PubMed]
- Eaton SL, Proudfoot C, Lillico SG, Skehel P, Kline RA, et al. (2019) CRISPR/Cas9 mediated generation of an ovine model for infantile neuronal ceroid lipofuscinosis (CLN1 disease). Sci Rep. 9(1):9891.
[Crossref] [Google Scholar] [PubMed]
- Gao Y, Wu H, Wang Y, Liu X, Chen L, et al. (2017) Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome Biol. 18(1):1-15.
[Crossref] [Google Scholar] [PubMed]
- Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157(6):1262-1278.
[Crossref] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol Res. 169:5429-5433.
[Crossref] [Google Scholar]
- Jiang F, Doudna JA (2017) CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 46:505-529.
[Crossref] [Google Scholar] [PubMed]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337(6096):816-821.
[Crossref] [Google Scholar]
- Lino CA, Harper JC, Carney JP, Timlin JA (2018) Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 25:1234-1257.
[Crossref] [Google Scholar] [PubMed]
- Menchaca A, Anegon I, Whitelaw CBA, Baldassarre H (2016) New insights and current tools for Genetically Engineered (GE) sheep and goats. Theriogenology. 86:160-169.
[Crossref] [Google Scholar] [PubMed]
- Menchaca A, Dos Santos-Neto PC, Mulet AP, Crispo M (2020) CRISPR in livestock: From editing to printing. Theriogenology. 150:247-254.
[Crossref] [Google Scholar] [PubMed]
- Moradpour M, Abdulah SNA (2020) CRISPR/dC as9 platforms in plants: Strategies and applications beyond genome editing. Plant Biotechnol J. 18:32-44.
[Crossref] [Google Scholar] [PubMed]
- Ni W, Qiao J, Hu S, Zhao X, Regouski M, et al. (2014) Efficient gene knockout in goats using CRISPR/Cas9 system. PloS One. 9:106718.
[Crossref] [Google Scholar] [PubMed]
Citation: Kumar MV, Kumar S (2023) The Overview of CRISPR Technology and its Application on Farm Animals. J Animal Sci Livest Prod. 7:41.
Copyright: © 2023 Mehra VK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.