Research Article - (2018) Volume 4, Issue 2
Joséphine Briand1,2,3,4,5, Arulraj Nadaradjane1,2,3,4,5, François M Vallette1,2,3,5,6 and Pierre-François Cartron1,2,3,4,5,6*
1Equipe Apoptose & Progression Tumorale, Centre de Recherche en Cancérologie et Immunologie Nantes Angers (CRCINA), INSERM U1232 Nantes, France
2Faculté de Médecine, Université de Nantes, France
3LaBCT, Institute de Cancérologie de l'Ouest, Saint Herblain, France
4Cancéropole Grand-Ouest, réseau Epigénétique (RepiCGO), France
5EpiSAVMEN Consortium (Région Pays de la Loire), France
6LabEX IGO, Université de Nantes, France
*Corresponding Author:
Pierre-François Cartron
CRCINA, INSERM U1232, Equipe Apoptose & Progression Tumorale
Centre de Recherche en Cancérologie et Immunologie Nantes Angers (CRCINA)
INSERM U1232 Nantes, France.
Tel: +33243484785
E-mail: pierre-francois.cartron@inserm.fr
Received date: March 28, 2018; Accepted date: April 17, 2018; Published date: April 26, 2018
Citation: Briand J, Nadaradjane A, Vallette FM, Cartron PF (2018) The TET2 Expression Level Correlates with a Short Relapse Time in Glioblastoma Multiforme. J Clin Epigenet 4:12. doi: 10.21767/2472-1158.100097
Background: Epigenetic changes play crucial roles in cancer initiation and progression and in tumor recurrence. Glioblastoma multiforme (GBM) is by far the most common and most malignant of the glial tumors, and its recurrence is ineluctable. Thus, anti-GBM therapy need for new treatments for recurrent GBM. We here in focused on identifying whether TET2 (an epigenetic players involving in the DNA demethylation process) as a putative target for the development of anti-GBM therapy.
Results: We observed that the expression level of TET2 at mRNA and protein levels is not associated with a prognosis value of survival in GBM patients. However, in a cohort of 10 GBM patients having received two surgical resections (rGBM), we noted that three in ten rGBM patients have an increase of TET2 expression between resection#1 and #2.
Conclusions: By observing an increase of TET2 expression between two surgical resections an despite the fact that expression level of TET2 is devoid of prognosis value of survival in GBM patients, our data provide a promising starting point for the use of TET2 inhibitors administrable in the subgroup of patients with recurrent GBM.
Keywords
DNA methylation; TET; TET2; Glioblastoma multiforme; TET inhibitors
Introduction
The Ten-Eleven Translocation-2 (TET2) is a member of the TET family proteins that include 2 other members: TET1 and TET3. These proteins catalyze the steps of the active DNA cytosinedemethylation via the conversion of 5-methylcytoisne (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxycytosine (5CaC) [1-3]. TET2 such as other TET proteins is an α-ketoglutarate- and Fe2+-dependent dioxygenase and the oncometabolite 2-hydroglutarate inhibits its activity [4].
In cancer, the TET2 mutational state is frequently associated with a prognosis value. However, TET2 mutations are more present in liquid tumors than in solid tumors. As example, TET2 mutations are rare in breast, ovarian, lung and brain cancers and observed with a prevalence of 6%-26% in MDS, or in 12-27% in adult AML [5]. In glioblastoma multiforme (GBM), 7 genetic alterations within TET2 (p.V218M, p.G355N, p.P363L, p.L1721W, p.P1723S, p.I1762V, p.H1778R), are described, but the presence of these alterations was not associated with a prognosis value [6]. Investigations performed with human samples and murine models suggest that TET2 deficiency does not promote the tumorigenesis but predisposes to the development of certain tumors such as MPN, MDS and lymphoma [7]. In other terms, these works suggest that TET2 could act as tumor suppressor gene [8-10]. However, TET2 could act as an oncogene by participating to the global DNA hypomethylation phenotype i.e. oncogenic phenotype. A recent article suggests that TET2 sustains immunosuppressive function of tumor-infiltrating myeloid cells to promote melanoma progression [11]. Thus, this last article supports the idea that TET2 could be a therapeutic target in certain cancers (other than hematologic cancer, certainly).
The role of TET proteins in glioblastoma multiforme is not fully documented. Two articles report the involvement of TET3 in GBM. Cui et al. suggest that TET3 acts as a tumor suppressor gene in glioblastoma stem cells [12]. Two articles report that TET3 is involved in the dynamic methylation/demethylation mechanism in GBM cells [13,14]. Takai et al. report that TET1-mediated production of 5-hydroxymethylcytosine (5hmC) is required for the tumorigenicity of glioblastoma cells [15]. The nuclear exclusion of TET1 is also reported as a source of DNA methylation and a loss of 5-hydroxymethylcytosine [16]. However, the large majority of articles investigating the role of TET1 in glioma is focused on low-grade glioma. A similar observation can be performed about TET2 [17,18]. Chen et al. reports that the modulation of TET2 expression affects the invasion potential and the proliferation rate of glioma cells [19]. Always in low-grade glioma, Guilhamon et al. report that TET2 interacts with EBF1 to promote a sequencespecific mechanism of DNA demethylation [20]. Thus, there is a lack of investigation on the role of TET2 on GBM. In order to try to overcome this deficiency, our present study investigated the putative association of TET2 with a prognosis value in the context of GBM and its dynamic expression following first resection and standard anti-GBM treatment.
Methods
Tumors samples
Tumors were collected from the “Réseau des tumorothèques du Canceropole Grand-Ouest/réseau Gliome”, the “Base clinicobiologique des Glioblastomes (n°BRIF: BB-0033-00093), CHU Angers, France" and the Biological Resource Center of University hospital of Angers. Tumor patient material and records (diagnosis, age, sex, date of death) were used with confidentiality in accordance with French laws and the recommendations of the French National Ethics Committee. In accordance with the regulations, all subjects signed a specific informed consent form for this biocollection, approved by an Ethics Committee, the French State Department for National Education, Higher Education and Research and the CNIL.
Protein analysis: ELISA
Proteins extracts were obtained by using RIPA Lysis and Extraction Buffer (Thermo Scientific, France) in accordance with the manufacturer’s instructions. TET2-ELISAs were performed according to the manufacturer’s instructions (MyBiosource, MBS9317739, USA).
Statistics
The log-rank test was used to test whether the difference between survival times of two groups is statistically different or not. The Kaplan-Meier survival curves were used to represent the probability of surviving in a given length of time of subgroups of patients. T-test is performed to estimate whether the difference between a parameter characterizing two groups is statistically different or not. Pearson’s correlation test is performed to estimate the significant character of a correlation between two parameters.
Results
The TET2mRNA level is not associated with a prognosis value
To investigate whether TET2mRNA can be used as a biomarker associated with a favorable survival, we first analyzed data available on “Betastais/REMBRANDT database” (https://www.betastasis.com/glioma/rembrandt/kaplan_meier_survival_curve/). Thus, we observed that the expression level of TET2mRNA in GBM is not associated with a prognosis value of survival, neither in term of overall survival (OS) (Figure 1).
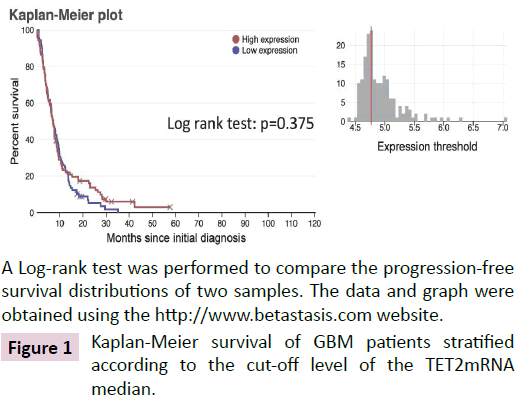
A Log-rank test was performed to compare the progression-free survival distributions of two samples. The data and graph were obtained using the https://www.betastasis.com website.
Figure 1: Kaplan-Meier survival of GBM patients stratified according to the cut-off level of the TET2mRNA median.
The TET2 expression level is not associated with a prognosis value
In a second approach, we have considered the TET2 protein expression level as a putative biomarker associated with a prognosis survival. TET2 expression level was analyzed in 31 GBM patients. The patients' characteristics of this cohort (cohort#1) are listed in (Table 1). ELISA was performed to calculate the TET2 expression level in GBM (Figure 2A). Our data indicates significant heterogeneity in the TET2 expression since a 11-fold increase was observed between the minimal value and the maximal value for TET2.
| Cohort#1 (n=31) | Characteristics | Patients | Log-rank test | |
|---|---|---|---|---|
| Age (years) Median (range) | 56[46;69] | p=0.30 | ||
| Gender | p=0.29 | |||
| Male | 20 | |||
| Female | 11 | |||
| Survival time (months) | ||||
| Median (range) | 14.1[4.6;42.6] | |||
| Extent of surgery | ||||
| Biopsie or Partial resection | 0 | |||
| Complete resection | 31 | |||
| Cohort#2 (n=10) | Age (years) Median (range) | 51[42;58] | np | |
| Gender | np | |||
| Male | 7 | |||
| Female | 3 | |||
| Survival time (months) | ||||
| Median (range) | 23.6[11;36.3] | |||
| Extent of surgery#1 and #2 | ||||
| Biopsie or Partial resection | 0/0 | |||
| Complete resection | 10 | |||
Table 1: Patients' characteristics of this cohort (cohort#1).
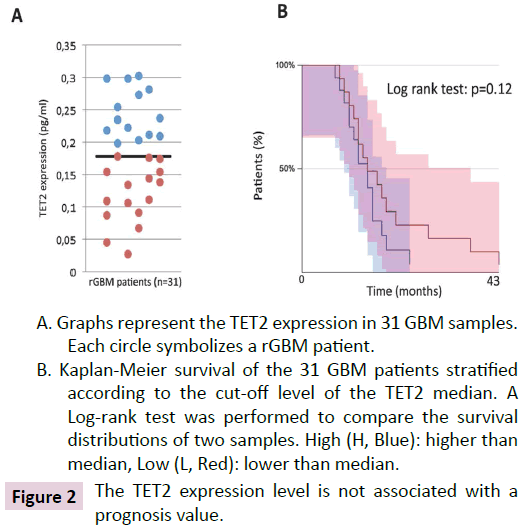
A. Graphs represent the TET2 expression in 31 GBM samples. Each circle symbolizes a rGBM patient.
B. Kaplan-Meier survival of the 31 GBM patients stratified according to the cut-off level of the TET2 median. A Log-rank test was performed to compare the survival distributions of two samples. High (H, Blue): higher than median, Low (L, Red): lower than median.
Figure 2: The TET2 expression level is not associated with a prognosis value.
Then, our cohort of 31 GBM samples has been divided in 2 subgroups using the median value as threshold. Survival curves were visualized in Kaplan-Meier plot. A log-rank test indicates a lack of difference between the overall survival of GBM patients having a high level of TET2 and those having a low level of TET2 (Figure 2B).
The TET2 expression level correlates with a short relapse time in recurrent GBM
We extended our study by asking if the TET2 expression could be associated with a short relapse time in recurrent GBM (rGBM). To address this question, TET2 expression level was analyzed by ELISA in resection#1 and #2 of 10 GBM patients having received two surgeries. The relapse time is here defined such as the months separating the both resections. The patients' characteristics of this cohort (cohort#2) are listed in Table 1. All GBM of this cohort are IDH1 wild-type and are primary GBM (i.e. that the patients are devoid of medical history with low grade glioma). Thus, we noted that three in ten rGBM patients have an increase of TET2 expression between resection#1 and #2 (Figure 3A). One in ten rGBM patients harbors a decrease of TET2 expression (Figure 3A). We next calculated the changes of TET2 expression between resection #1 and #2 (#TET2=TET2Resection#2-TET2Resection#1). Graph and Pearson’s correlation test indicate that the changes of TET2 expression between resection#1 and #2 was correlated with relapse time (p=0.0377) (Figure 3B).
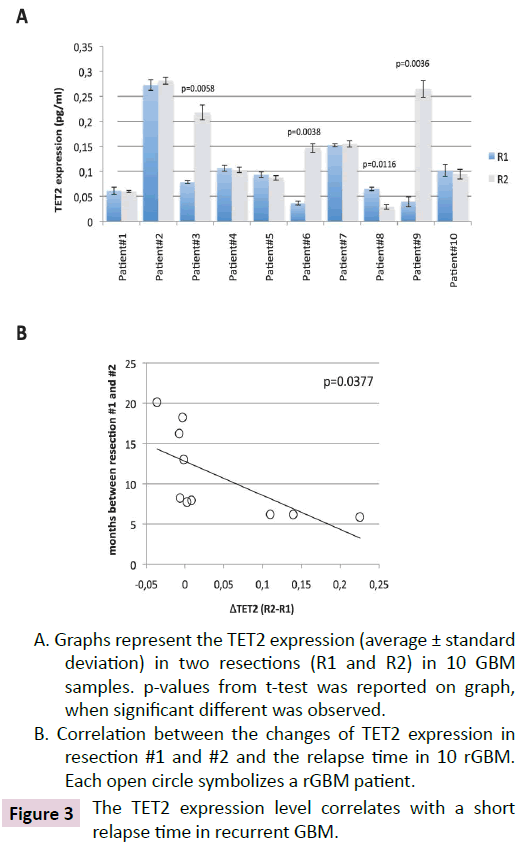
A. Graphs represent the TET2 expression (average ± standard deviation) in two resections (R1 and R2) in 10 GBM samples. p-values from t-test was reported on graph, when significant different was observed.
B. Correlation between the changes of TET2 expression in resection #1 and #2 and the relapse time in 10 rGBM. Each open circle symbolizes a rGBM patient.
Figure 3: The TET2 expression level correlates with a short relapse time in recurrent GBM.
Discussion
Glioblastoma multiforme (GBM) is the most common and aggressive primary brain cancer. Despite the gain of survival associated with the use of a standard of care treatment composed by a maximal safe surgery followed by adjuvant chemoradiotherapy and adjuvant chemotherapy, recurrence is an inevitable event. Besides, literature reports that GBM patients underwent repeat resections with a median time between resections of 6-9 months [21,22]. On contrary to the first line of treatment, the treatment of recurrent GBMs (rGBMs) poses a great challenge since no well-defined recommendation of therapy is suggested. This point finds its echo in the multiples clinical trials performed about the rGBM treatment and with a large panel of drugs including: PARP inhibitor (Veliparinb, ClinicalTrials. gov Identifier: NCT01026493), vEGF inhibitor (ClinicalTrials. gov Identifier: NCT01474239), or EGFr inhibitor (ClinicalTrials. gov Identifier: NCT01310855). Several epidrugs such as HDAC inhibitor (ClinicalTrials.gov Identifier: NCT01110876) and BET inhibitor (OTX015 ClinicalTrials.gov Identifier: NCT02296476) have also been investigated for the treatment of rGBM [23]. The data included in our study suggests that TET2 expression could be used as a putative biomarker and/or target for the development of precision and personalized medicine for the treatment of rGBM, but not for the initial GBM treatment. Despite the limitation of this initial study (low number of rGBM), our data constitute a promising starting point for the design of TET2 inhibitors administrable at patients with rGBM i.e. for patients having a strong medical need.
The observation of a correlation between an elevated TET2 expression and a short time of relapse also asks the question of the role played by TET2 in the GBM recurrence. Thus, several hypotheses can be formulated from this observation: is the TET2 overexpression an actor of the GBM recurrent?; is the TET2 overexpression a biomarker associated with the acquired resistance of the standard anti-GBM treatment? Is the TET2 overexpression a consequence of the GBM recurrence?....and others. Investigations replying at these questions are ongoing in our lab.
Conclusion
Finally, our data reinforce the need for the development of TET2 inhibitors. On contrary to the other epigenetic players, the epidrug pipeline is, to date, poor in TET inhibitors and more particularly in TET2 inhibitors. Several points can explain this relative lack. Thus, the fact that TET2 protein was later characterized as connected to cancer than others epigenetic players could explain the lack in TET2 inhibitors of the epidrug pipeline [24,25]. The focus on TET2 mutations and not on the TET2 expression level could be another possible explanation. The description of TET2 as both tumor suppressor gene and oncogene also appears be a drag for the development of TET2 inhibitors. Literature reports that DMOG (an antagonist of α-ketoglutarate cofactor and inhibitor for HIF prolylhydroxylase) and 2-hydroxyglutarate can be used to inhibit TET2 [26,4]. However, these molecules having a large spectrum of selectivity, their use such as selective TET2 inhibitors can be discussed.
In conclusion, our data and the current lack of selectivity of TET2 inhibitors support the research for a better understanding of the role played by TET2 in rGBM and for the design/development of highly selective TET2 inhibitors.
Declarations
Acknowledgements
JB was supported by a fellowship from EpiSAVMEN/REGION PAYS DE LA LOIRE and “EN AVANT LA VIE”, a French association that fights against glioma. We thank to Dr P Menei and G Soulard for coordinating the patient sample procurement from BCB-G.
Funding
This work was supported by grants from the LIGUE NATIONALE CONTRE LE CANCER, “Comité InterRégional Grand Ouest, département de Loire-Atlantique, Vendée et Morbihan (AO2015/ Subvention2016)”.
Authors’ Contributions
PFC designed and coordinated the project.
JB, AN and PFC performed all experiments.
FMV and PFC interpreted and discussed the data.
PFC wrote the first version of the manuscript and all authors reviewed and approved it.
Conflicting Interests
The authors declare that they have no conflicting interests.