Research Article - (2017) Volume 1, Issue 1
Jean-Claude Perez*
Retired Interdisciplinary Researcher (IBM emeritus), 7 Avenue de Terre-rouge, F33127 Martignas, Bordeaux Metropole, France
*Corresponding Author:
Jean-Claude Perez
PhD Maths and Computer Science
Retired Interdisciplinary Researcher (IBM emeritus)
7 Avenue de Terre-rouge F33127 Martignas
Bordeaux Metropole, France
Tel: 330781181112
E-mail: jeanclaudeperez2@gmail.com
Received Date: September 27, 2017; Accepted Date: October 06, 2017; Published Date: October 13, 2017
Citation: Perez JC (2017) Towards a Universal Law Controlling All Human Cancer Chromosome LOH Deletions, Perspectives in Prostate and Breast Cancers Screening. Res J Oncol. Vol.1 No.1: 3
Background: Global analysis of 3 human genomes of increasing levels of evolution (Neanderthal/Sapiens Build34/Sapiens hg38) reveals 2 levels of numerical constraints controlling, structuring and optimizing these genome's DNA sequences. A global constraint - called "HGO" for "Human Genome Optimum" - optimizes the genome at its global scale. The same operator applied to each of the 24 individual chromosomes reveals a hierarchical structure of these 24 chromosomes. Methods: We analyze how this HGO genomic optimum is perturbed by hundred single or multiple LOH (Loss of Heterozygosity) deletions relating to different chromosomes and cancers. Results: The generic law highlighted is stated as follows "When an LOH deletion affects a chromosome upstream of the HGO point (chromosomes 4 13). In the chromosomal spectrum, this deletion degrades the genomic optimum of the cancer genome. When an LOH deletion affects a chromosome downstream of the HGO point (chromosomes 19 22) in the chromosomal spectrum, this deletion improves the genomic optimum of the cancer genome. The exhaustive analysis of the 240 LOHs for the following 6 cases: Chromosome 13 (breast cancer), chromosome 5 (breast cancer), chromosome 10 (glioblastoma cancer), chromosome 1 (colorectal cancer), chromosome 1 (neuroblastoma cancer) and chromosome 16 (prostate cancer) obey this law in 227 cases and do not obey this law for 13 cases (success rate of our law=94.58%). In this article we will detail this type of analysis on 153 LOH relating to breast and prostate tumors affecting respectively chromosome 13, chromosome 5 (breast) and chromosome 16 (prostate). In this detailed study, the HGO law described here is verified in 143 cases out of 153 or 93.46% of favorable cases. Conclusion: The main application of this fundamental discovery will be the genomic characterization and classification of tumors, making it possible to predict the dangerousness and even the pathogenicity.
Keywords
Human genome; Cancer; Loss of Heterozygosity; Biomathematics; Evolution
Introduction
Thanks to the CRISPR (Clustered regularly interspaced short palindromic repeats) technology, it is now possible to locally modify the genomes, and particularly the human genome [1]. Almost simultaneously, the fractal and global structures of the human genome were demonstrated [2]. In such a context, apart from ethical questions, can a local technology as powerful as CRISPR be applied, ignoring its possible effect on the possible global and long-range equilibria and balancing at the chromosome scale or even the entire genome scale? For more than 25 years, we have been looking for possible global, even numerical, structures that would organize DNA, genes, chromosomes and even whole genomes [3-6]. Curiously, the method of numerical analysis of whole genomes as well as the discovery of a global law of impact of the LOH deletions associated with tumors that will be the subject of this article may seem to the reader too simple or even "naive". In effect, these laws are based on a wellknown type of analysis that could be thought to have explored all aspects: the analysis of GC/TA ratios. It has long been known that such ratios [7] characterize the diversity of chromosomes. Under the name of "isochores," Giorgio Bernardi has extensively explored its richness and diversity [8,9]. We have already demonstrated a numerical structure at the scale of each human chromosome as well as on the whole genome [10-13]. In [10] we have already highlighted this numerical value of 0.6909830056, the HGO in this article: it controls the population of triplet codons analyzing single stranded DNA sequence from the whole human genome. However, it is only by deepening the notion of "fractal periodicity", outlined in [14], and we will highlight in various articles in preparation [15] that we have re-discovered the major role that this GC/TA ratio at the whole chromosome and whole genome scales. Particularly, in 2015, we have demonstrated [14] how a global structure organizes any DNA sequence. This numerical organization is based on the atomic masses of the "G" bases of the double stranded DNA sequence, therefore on the bases C and G of the single stranded DNA sequence. Here is another reason sufficient to consider the relations and proportions between bases C+G on the one hand, and T+A on the other hand. Comparing the three genomes of reference of Neanderthal [15], Sapiens Build 34 of 2003 [16] and Sapiens hg38 of 2013 [17], we will demonstrate in [15] the evidence of "Fractals periods" and "resonance periods" characterizing each of the 24 human chromosomes. It is for these simple reasons that we have decided to focus on these C+G/T+A ratios at the scale of each of the 24 human chromosomes as well as on the whole genome analyzing the 46 chromosomes single stranded DNA sequences.
Materials and Methods
Analyzed whole human genomes
We analyzed completely and systematically each of the 24 chromosomes of each of the following three reference genomes:
Neanderthal genome: Neanderthal genome (2014) [15] https://www.nature.com/nature/journal/v505/n7481/full/nature12886.html Sapiens Build 34 (2003) human genome [16]: Sapiens Build34 (2003) genome https://www.nature.com/nature/journal/v431/n7011/full/nature03001.html Sapiens HG38 (2013) human genome [17]: Sapiens HG38 (2013) genome https://www.ncbi.nlm.nih.gov/grc/human
Computing the HGOs
Let us now distinguish the three types of HGO that will be discussed
Theoretical HGO (tHGO): tHGO=(3-PHI) ÷ 2=0.6909830056, where PHI is the Golden Ratio PHI=1.618033989.
Reference woman HGO (rwHGO): rwHGO=0.6913477936, error (tHGO-rwHGO)=0.6909830056-0.6913477936=0.0003647879784 and Reference man HGO (rmHGO): rmHGO=0.6922864236, error (tHGO-rmHGO)=0.6909830056-0.6922864236=0.001303417973.
HGO (LOH) is considered as a function: With LOH the deletion disturbing the computed CG/TA whole human genomic ratio and « n » the LOH partially deleted chromosome and « chrs » all chromosomes from 1 to 22 which are different with « n ». HGO is computed by compiling C+G and T+A nucleotides populations from each 46 chromosomes DNA single strand.
Example: HGOwoman (LOH chr n)=[(C+G population from 46 chromosomes strand1)+(C+G population from 46 chromosomes strand1)]/[(T+A population from 46 chromosomes strand1)+(T+A population from 46 chromosomes strand1)]. Then: HGOwoman (LOH chr n)=[(sum C+G single strand 1 to 22 chromosomes except chrn)+(sum C+G chrn)+(sum C+G chrX)+(sum C+G single strand 1 to 22 chromosomes except chrn)+(sum C+G chr LOH n)+(sum C+G chrX)]/[(sum T+A single strand 1 to 22 chromosomes except chrn)+(sum T+A chrn)+(sum T+A chrX)+(sum T+A single strand 1 to 22 chromosomes except chrn)+(sum T+A chr LOH n)+(sum T+A chrX)]. HGOman (LOH chr n)=[(sum C+G single strand 1 to 22 chromosomes except chrn)+(sum C+G chrn)+(sum C+G chrX)+(sum C+G single strand 1 to 22 chromosomes except chrn) +(sum C+G chr LOH n)+(sum C+G chrY)]/[(sum T+A single strand 1 to 22 chromosomes except chrn)+(sum T+A chrn)+(sum T+A chrX) +(sum T+A single strand 1 to 22 chromosomes except chrn)+(sum T+A chr LOH n)+(sum T+A chrY)] [18,19].
Results
In all that follows, the general methodology will be as follows: we calculate, for the 46 chromosomes constituting each genome studied, only the single-stranded DNA sequences. In these sequences, we count the relative populations of bases T+A on the one hand, and C+G on the other hand.
HGo of the 3 whole genomes: Neanderthal, Sapiens Build34 and Sapiens hg38
The three genomes we compare here are differentiated on the one hand by their respective evolution levels, on the other hand by the sample of individual genomes of which they form the syntheses, and finally by the precision of the sequencing of DNA.
For example, this work by the Craig Venter team demonstrates this level of dispersion between more than 10,000 individual genomes and reference genomes such as HG28 [20].
The 3 analysed genomes are:
• Neanderthal genome (2014) [15]
• Sapiens Build34 (2003) genome [16]
• Sapiens hg38 (2013) genome [17]
The methods of computing the different HGOs are given in "supporting information part 1".
The detailed data related to the 3 whole genomes shows the various distances and errors between real computed HGOs for each genome and theoretical HGO optimum value= 0.6909830055.
Particularly, it is found that the 3 HGOs calculated for the respective 3 genomes of Neanderthal, Sapiens (2003 Build34 and 2013 hg38 Sapiens) are very close to the ideal theoretical optimal HGO=0.6909830056 (99.67% for the least optimal genome).
It is also observed that female genomes (XX) are more optimal than male genomes (XY).
On the other hand, the genomes of Neanderthal and Sapiens (Build34 of 2003) have very close optimization levels. We believe these results from the fact that the precisions of their respective DNA sequencing are similar. On the contrary, the hg38 genomes of 2013 show the most optimal levels, this is most certainly due to the deeper quality of their DNA sequencing. Figure 1 summarizes HGO results for these 3 human genomes of varying levels of evolution.
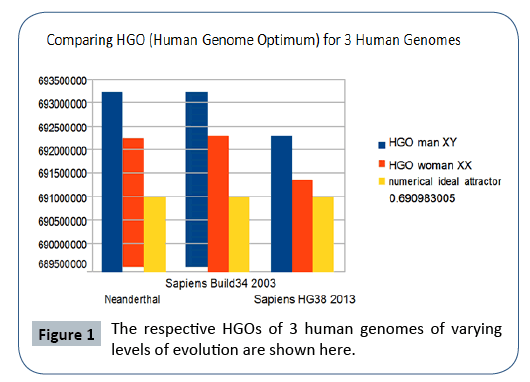
Figure 1: The respective HGOs of 3 human genomes of varying levels of evolution are shown here.
Finally, we note that it is the downstream region (Figure 2) that contributes the most to the superiority of optimality of sapiens hg38 compared to sapiens Build34. However, a more detailed analysis of the upstream and downstream regions shows that when passing from sapiens Build34 2003 to hg38 2013, the notorious optimization obeys the global strategy law of the LOH. Also of note is that 20 out of 24 chromosomes are OK, with the exception of chromosomes 13, X, Y, and 1. The law presented here states that there is reduction of optimality for upstream chromosomes and improvement of optimality for downstream regions.
It is therefore remarkable that this is the same and only law which seems to simultaneously control two domains as different as: 1) the global strategy of LOH deletions of tumors on the one hand and 2) the difference of evolution of the same genome following two increasing precision levels of DNA sequencing (Build34 2003 and hg38 2013) on the other.
Recall the single stranded C+G, T+A and individual chromosome HGO values:
In Table 1, we have sorted the 24 chromosomes by increasing values of CG/TA ratios in the 3 cases of compared genomes.
Table 1 CG values, TA values and CG/TA ratio for each sapiens HG38 individual chromosome.
| Chromosome | C+G | T+A | (C+G)/(T+A) |
|---|---|---|---|
| Chromosomes upstream HGO point=(3-Phi)÷2=0.6909830056 | |||
| 4 | 72568001 | 117184666 | 0.6192619178 |
| 13 | 37772797 | 60210328 | 0.627347 |
| 5 | 37772797 | 60210328 | 0.627347471 |
| X | 71611274 | 109654104 | 0.6530651511 |
| 6 | 61221521 | 93671508 | 0.6535767632 |
| 3 | 67360020 | 102718502 | 0.6557729979 |
| 18 | 78577742 | 119522393 | 0.6574311309 |
| Y | 31856106 | 48233499 | 0.6604560453 |
| 8 | 10572683 | 15842360 | 0.66736793 |
| 2 | 58133960 | 86634176 | 0.6710280248 |
| 7 | 96769083 | 143779145 | 0.6730397722 |
| 12 | 64696843 | 94273288 | 0.686269084 |
| 14 | 54275482 | 78862334 | 0.6882307338 |
| Chromosomes downstream HGO point=(3-Phi)÷2=0.6909830056 | |||
| 21 | 16411625 | 23676994 | 0.693146478 |
| 9 | 50270473 | 71520077 | 0.70288617 |
| 11 | 55885058 | 78648684 | 0.7105657102 |
| 10 | 55359481 | 77903481 | 0.7106162689 |
| 1 | 96166571 | 134314441 | 0.7159808751 |
| 15 | 35578844 | 49062481 | 0.7251741713 |
| 20 | 28010605 | 35933652 | 0.7795089962 |
| 16 | 36472718 | 45333225 | 0.8045471726 |
| 17 | 37575444 | 45344760 | 0.8286612169 |
| 22 | 18406838 | 20752939 | 0.8869509037 |
| 19 | 28015712 | 30425046 | 0.9208108346 |
HGO spectral hierarchy of the 24 human chromosomes
The following Figures 2 and 3 illustrate the hierarchical spectrum of the individual HGOs of each of the 24 chromosomes for each of the three genomes analyzed. It should be noted that the upstream/downstream tipping point lies between chromosomes 14 and 21, which is closely related to the probable mechanisms explaining trisomy21 (whose disorders involve precisely these two chromosomes).
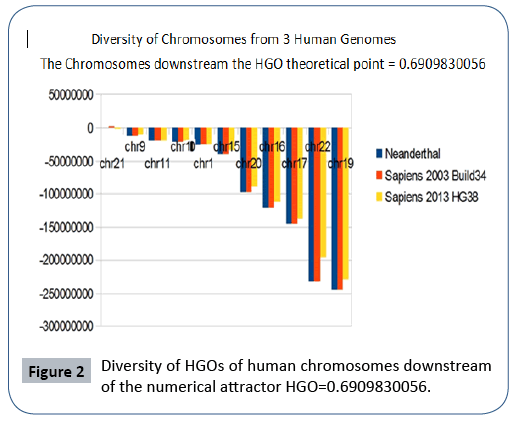
Figure 2: Diversity of HGOs of human chromosomes downstream of the numerical attractor HGO=0.6909830056.
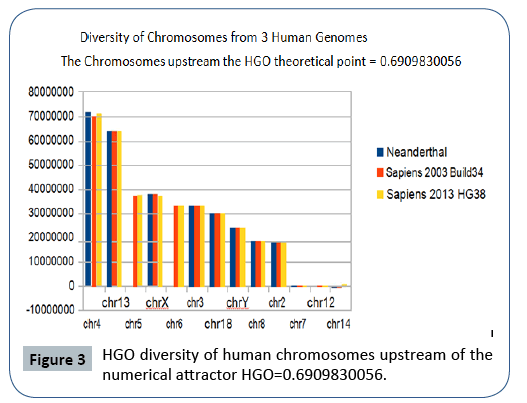
Figure 3: HGO diversity of human chromosomes upstream of the numerical attractor HGO=0.6909830056.
It then reveals a hierarchical classification scale of 24 chromosomes ranging from 1/Phi (chromosome4) to 3/2 Phi (chromosome 19).
HGO disturbed by 240 LOH deletions related to 5 different chromosomes and 5 different tumors.
After demonstrating how 3 human genomes with increasing levels of sequencing quality and evolution accuracy seem to "fit" this theoretical value of HGO, we will now analyze how small mutations on one of the chromosomes can "disrupt" this HGO. For this purpose there are a very large number of publications associating such nucleotide regions with deletions of the chromosomal DNA of different tumor cells.
We analyzed the Loss of Heterozygosity associated with 240 cases published in [21-26]. These 240 cases relate successively to the Breast [21,22], Glioblastoma [23], Colorectal [24], Neuroblastoma [21], and Prostate [23] cancers. They successively affect the chromosomes 13, 5, 10, 1 and 16.
This exhaustive analysis will then reveal the following generic law: "When an LOH deletion affects a chromosome upstream of the HGO point, in the chromosomal spectrum, this deletion degrades the genomic optimum of the cancer genome.
When an LOH deletion affects a chromosome downstream of the HGO point, in the chromosomal spectrum, this deletion improves the genomic optimum of the cancer genome.
The full details of these analyses are available in Table 2, below summarizing these results.
Table 2 Table of synthesis of 240 LOH relating to 5 types of cancer and 5 different chromosomes, the shaded areas represent those that comply with the HGO law, i.e., 227 cases out of 240.
| Cancer type | Chromosome LOH deletions |
LOH deletions decreasing HGO (Human Genome Optimum) | LOH deletions increasing HGO (Human Genome Optimum) | Total number of LOH | % law is OK |
|---|---|---|---|---|---|
| Upstream HGO | |||||
| Breast | Chromosome 13 | 36 | 4 | 40 | |
| Breast | Chromosome 5 | 37 | 5 | 42 | |
| Downstream HGO | |||||
| Glioblastoma | Chromosome 10 | 1 | 14 | 15 | |
| Colorectal | Chromosome 1 | 0 | 33 | 33 | |
| Neuroblastoma | Chromosome 1 | 2 | 36 | 38 | |
| Prostate | Chromosome 16 | 1 | 71 | 72 | |
| Total | 77 | 163 | 240 | ||
| LOH deletions respecting the HGO Law | LOH deletions unrespecting the HGO Law | Total number of LOH | % law is OK | ||
| Total | 227 | 13 | 240 | 94.58% | |
In Table 2 the shaded areas represent cases of LOH complying with the generic rule, while the unshaded areas represent cases of LOH that do not comply with this rule. In total, 227 cases respect the rule and only 13 cases of LOH do not respect it, a success rate of this law equal to 94.58%. In Bold we represented the 153 Breast and prostate LOH deletions cases detailed in this article.
Global graphical analysis of the 240 LOH tumor cases: In Figures 4-7 the 4 images below present an overall analysis of the HGOs resulting from the 240 cases of LOH deletions associated with 5 types of cancers and 5 distinct chromosomes.
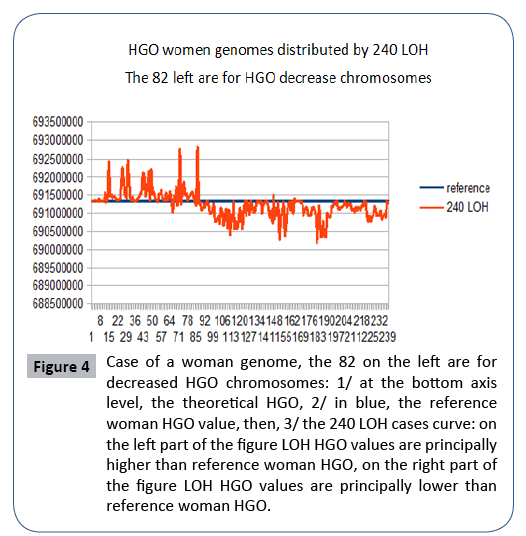
Figure 4: Case of a woman genome, the 82 on the left are for decreased HGO chromosomes: 1/ at the bottom axis level, the theoretical HGO, 2/ in blue, the reference woman HGO value, then, 3/ the 240 LOH cases curve: on the left part of the figure LOH HGO values are principally higher than reference woman HGO, on the right part of the figure LOH HGO values are principally lower than reference woman HGO.
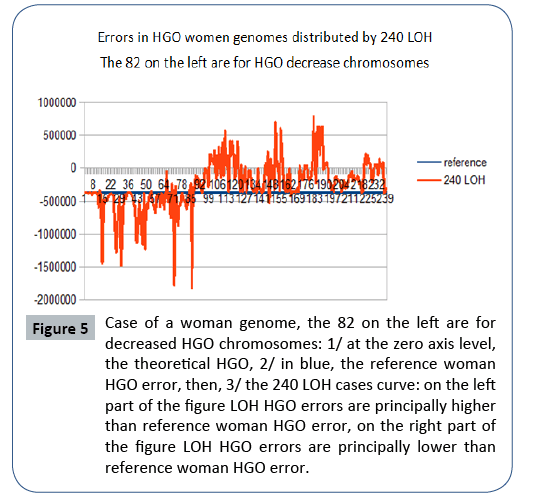
Figure 5: Case of a woman genome, the 82 on the left are for decreased HGO chromosomes: 1/ at the zero axis level, the theoretical HGO, 2/ in blue, the reference woman HGO error, then, 3/ the 240 LOH cases curve: on the left part of the figure LOH HGO errors are principally higher than reference woman HGO error, on the right part of the figure LOH HGO errors are principally lower than reference woman HGO error.
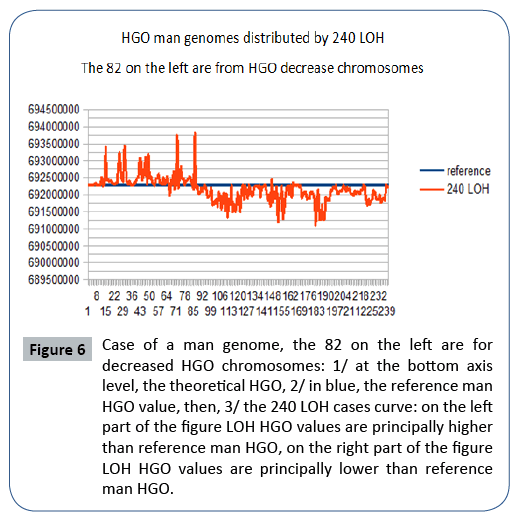
Figure 6: Case of a man genome, the 82 on the left are for decreased HGO chromosomes: 1/ at the bottom axis level, the theoretical HGO, 2/ in blue, the reference man HGO value, then, 3/ the 240 LOH cases curve: on the left part of the figure LOH HGO values are principally higher than reference man HGO, on the right part of the figure LOH HGO values are principally lower than reference man HGO.
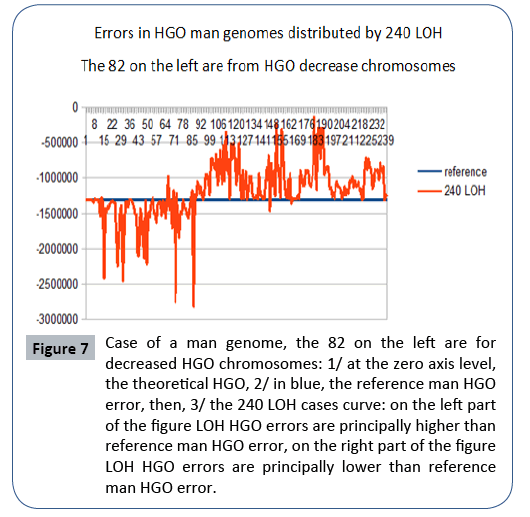
Figure 7: Case of a man genome, the 82 on the left are for decreased HGO chromosomes: 1/ at the zero axis level, the theoretical HGO, 2/ in blue, the reference man HGO error, then, 3/ the 240 LOH cases curve: on the left part of the figure LOH HGO errors are principally higher than reference man HGO error, on the right part of the figure LOH HGO errors are principally lower than reference man HGO error.
For this purpose we recall how to distinguish 3 types of HGO: the theoretical HGO:(3-Phi)÷2=0.6909830055, where Phi is the golden ratio Phi=1.618033.
The 2 reference HGOs relating to the respective hg38 reference genomes of the female (0.691347793) and man (0.692286423).
The 240 HGOs relating to each of the 240 chromosomes having ubi LOH deletions, On the other hand, the chromosomes are presented in these 4 figures according to the order sequence of the hierarchical spectrum shown in Figures 2 and 3 above.
Consequently, these 240 cases are presented in the order of upstream towards downstream with respect to the theoretical HGO.
In particular, this is the case for the first 82 LOHs for breast tumors affecting chromosomes13 and 5, both of which are upstream (Figure 3) on the contrary, all remaining LOHs will be downstream (Figure 2) of the theoretical HGO point, thus downstream.
Thus, Figures 4 and 6 will synthesize the relative positions of all these HGOs. Similarly, the errors or distances (Figures 5 and 7) will also be calculated relative to the respective distances between HGO of the 240 cases of LOH deletions, or of the reference HGOs, with respect to the HGO theoretical.
These 4 graphs of Figures 4 to 7 summarize the relevance of the generic law in visual form. Indeed, the fact of having ordered the chromosomes and their associated LOHs according to the classification of the 24 chromosomes of Figures 2 and 3 has helped to highlight the statistical reality of this law.
Results
We have successively demonstrated that, in the Human Genome Evolution and in the precision of sequencing of the genomes, these genomes seemed to "seek" a sort of numerical optimum: HGO (Figure 1).
Then, we demonstrated how the LOH deletions affecting the tumor cell chromosomes obeyed a generic law based on the chromosome hierarchization illustrated in Figures 2 and 3.
However, an important open-ended question: do these LOH deletions affecting the HGO optimum have any potential significance for the pathogenicity and aggressiveness of these tumors?
This is what we are going to demonstrate now in 3 analyses of 153 Breast and Prostate LOH deletions tumors.
First analysis: 40 breast tumors LOH in chromosome 13
Basic data: The article supporting our analysis.
We analyze here 40 LOH associated with Breast cancer of chromosome 13 from the publication (19), of the 46 LOH cases published, only 40 cases of exploitable LOH were identified. Indeed some LOH are reduced to a single marker that is to say approximately 200000bp, which is too insignificant. For example, the first of the markers in this study is D13S1695: chr13: 32,849,425-33,049,658 200,234 bp. However, such a length of 200234 bp is too insignificant.
Example of LOH effect on the HGO, the case of a single LOH deletion in chromosome 13 breast tumor.
We therefore retain only the LOHs corresponding to regions delimited by 2 markers (for example the first LOH referenced in Table 3), as well as regions comprising at least 2 markers alone (for example the LOH referenced D13S1694 D13S267 in Table 3).
Table 3 Computing HGO ratio disturbed by a sample LOH on chr13 breast tumor.
| LOH references : number/ref number in published figure/ Markers Id. |
Deleted LOH regions | Woman genome XX | Man genome XY | ||||
|---|---|---|---|---|---|---|---|
| Reference Human Genome (HG38 2013) | Reference HGO 0.69134779 |
Error ideal HGO – reference HGO -0.00036 |
Reference HGO 0.69228642 |
Error ideal HGO – reference HGO -0.00130341 |
|||
| Region A | |||||||
| 2 4 D13S1694 D13S267 |
33097286 33790250 | 0.69135215 | -0.00036915 | 0 | 0.69229100 | 0.00130799 | 0 |
The chromosome support of our analysis: chr13:1- 114,364,328 114,364,328 bp [26]
The supporting (Figure 8) of our analysis:
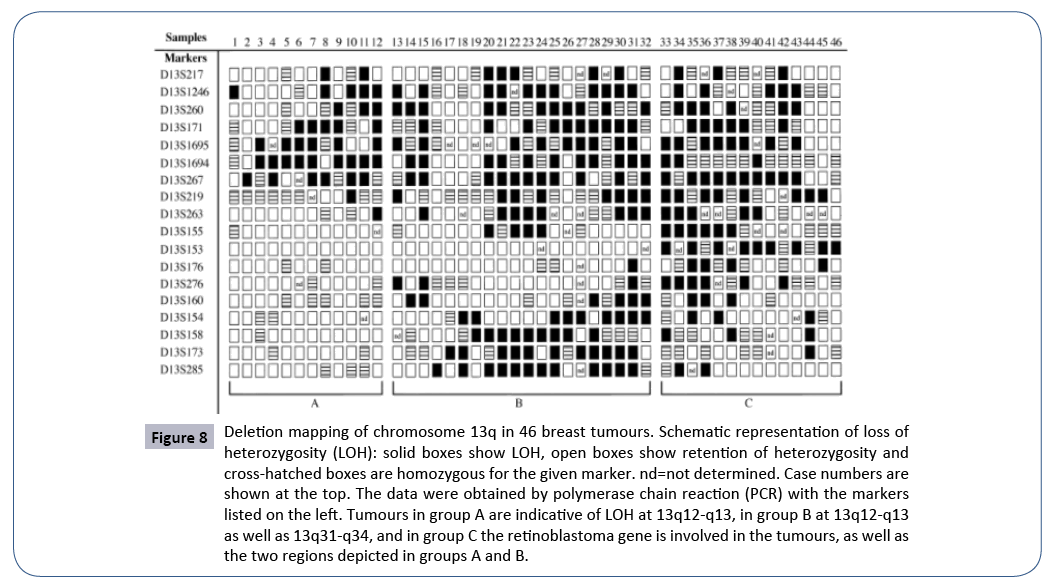
Figure 8: Deletion mapping of chromosome 13q in 46 breast tumours. Schematic representation of loss of heterozygosity (LOH): solid boxes show LOH, open boxes show retention of heterozygosity and cross-hatched boxes are homozygous for the given marker. nd=not determined. Case numbers are shown at the top. The data were obtained by polymerase chain reaction (PCR) with the markers listed on the left. Tumours in group A are indicative of LOH at 13q12-q13, in group B at 13q12-q13 as well as 13q31-q34, and in group C the retinoblastoma gene is involved in the tumours, as well as the two regions depicted in groups A and B.
Figure 8 the case analysed from Figure 1 in publication [19].
To Conclude: Recall that (Table 4) chromosome 13 is upstream with respect to the tipping point HGO (Figure 2). Thus, according to our "global strategy law of LOH by chromosomes", the majority of LOH "should" degrade the value of the HGO: in effect, we observe that 36 out of 40 cases degrade the HGO.
Table 4 Detail of 40 breast tumors LOH deletions of chromosome 13.
| LOH references : number / ref number in published figure / Markers Id. |
Deleted LOH regions | Woman genome XX | Man genome XY | ||||
|---|---|---|---|---|---|---|---|
| Reference Human Genome (HG38 2013) | Reference HGO 0.69134779 |
Error ideal HGO – reference HGO -0.00036478 |
OK | Reference HGO 0.69228642 |
Error ideal HGO – reference HGO -0.00130341 |
OK | |
| Region A | |||||||
| 1 3 D13S1695 D13S1694 |
32849425 33297510 | 0.69134714 | -0.00036413 | 1 | 0.69228583 | -0.00130282 | 1 |
| 2 4 D13S1694 D13S267 |
33097286 33790250 | 0.69135215 | -0.00036915 | 0 | 0.69229100 | -0.00130799 | 0 |
| 3 6 D13S171 D13S1694 |
32579775 33297510 | 0.69135118 | -0.00036817 | 0 | 0.69229001 | -0.00130700 | 0 |
| 4 7 D13S171 D13S267 |
32579775 33790250 | 0.69135563 | -0.00037262 | 0 | 0.69229464 | -0.00131163 | 0 |
| 5 8 D13S217 D13S1246 D13S171 D13S267 |
28697654 30631622 32579775 32780083 33589982 33790250 |
0.69134338 | -0.00036038 | 1 | 0.69228229 | -0.00129929 | 1 |
| 6 9 D13S260 D13S1694 | 31762621 33297510 | 0.69135713 | -0.00037412 | 0 | 0.69229623 | -0.00131323 | 0 |
| 7 10 D13S1246 D13S1694 D13S219 |
30431300 30631622 33097286 36684592 |
0.69138996 | -0.00040695 | 0 | 0.69233021 | -0.00134721 | 0 |
| 8 11 D13S217 D13S260 D13S1694 D13S267 |
28697654 31962968 33097286 33790250 |
0.69133825 | -0.00035524 | 1 | 0.69227730 | -0.00129430 | 1 |
| 9 12 D13S1246 D13S1694 D13S263 |
30431300 33297510 41406784 41607094 |
0.69134803 | -0.00036503 | 0 | 0.69228718 | -0.00130417 | 0 |
| Region B | |||||||
| 10 13 D13S1246 D13S260 D13S1695 D13S219 D13S276 |
30431300 31962968 32849425 33049658 36484263 36684592 68335674 68536043 |
0.69135049 | -0.00036748 | 0 | 0.69228953 | -0.00130653 | 0 |
| 11 14 D13S260 D13S1694 D13S267 D13S160 |
31762621 31962968 33097286 33790250 78504466 78704740 |
0.69135525 | -0.00037225 | 0 | 0.69229424 | -0.00131123 | 0 |
| 12 15 D13S1246 D13S267 D13S263 D13S276 D13S160 |
30431300 33790250 41406784 41607094 68335674 78704740 |
0.69157604 | -0.00059304 | 0 | 0.69252240 | -0.00153939 | 0 |
| 13 18 D13S154 D13S173 D13S285 |
95510028 95710383 107054541 112241249 |
0.69131884 | -0.00033583 | 1 | 0.69225765 | -0.00127465 | 1 |
| 14 19 D13S154 D13S158 |
95510028 103424175 | 0.69136848 | -0.00038548 | 0 | 0.69230891 | -0.00132590 | 0 |
| 15 20 D13S217 D13S171 D13S1694 D13S267 D13S155 D13S158 D13S285 |
28697654 32780083 33097286 33790250 53494781 53695047 10322447 103424175 112041068 112241249 |
0.69242518 | -0.00144217 | 0 | 0.69339874 | -0.00241573 | 0 |
| 16 21 D13S217 D13S260 D13S1694 D13S263 D13S158 D13S285 |
28697654 31962968 33097286 41607094 103224047 112241249 |
0.69146697 | -0.00048396 | 0 | 0.69241186 | -0.00142885 | 0 |
| 17 22 D13S217 D13S1695 D13S267 D13S155 D13S158 D13S285 |
28697654 28898018 32849425 33049658 33589982 53695047 103224047 12241249 |
0.69151941 | -0.00053641 | 0 | 0.69246701 | -0.00148400 | 0 |
| 18 23 D13S1246 D13S171 D13S1694 D13S267 D13S263 D13S155 D13S158 D13S285 |
30431300 30631622 32579775 32780083 33097286 33790250 41406784 53695047 103224047 112241249 |
0.69143522 | -0.00045222 | 0 | 0.69237962 | -0.00139662 | 0 |
| 19 24 D13S1246 D13S260 D13S1695 D13S267 D13S155 D13S158 D13S285 |
30431300 31962968 32849425 33049658 33589982 53695047 103224047 103424175 112041068 112241249 |
0.69147014 | -0.00048713 | 0 | 0.69241536 | -0.00143235 | 0 |
| 20 25 D13S1246 D13S171 D13S1694 D13S267 D13S154 D13S285 |
30431300 32780083 33097286 33790250 95510028 112241249 |
0.69141228 | -0.00042928 | 0 | 0.69235570 | -0.00137270 | 0 |
| 21 26 D13S260 D13S1695 D13S154 D13S158 |
31762621 33049658 95510028 103424175 |
0.69137794 | -0.00039494 | 0 | 0.69231881 | -0.00133580 | 0 |
| 22 27 D13S260 D13S267 D13S154 D13S173 |
31762621 33790250 95510028 95710383 107054541 107254838 |
0.69136439 | -0.00038139 | 0 | 0.69230382 | -0.00132081 | 0 |
| 23 28 D13S217 D13S1246 D13S171 D13S1694 D13S160 D13S158 D13S285 |
28697654 30631622 32579775 33297510 78504466 78704740 103224047 112241249 |
0.69138262 | -0.00039962 | 0 | 0.69232403 | -0.00134102 | 0 |
| 24 29 D13S1246 D13S171 D13S219 D13S154 D13S158 D13S285 |
30431300 32780083 36484263 36684592 95510028 95710383 103224047 112241249 |
0.69139238 | -0.00040937 | 0 | 0.69233399 | -0.00135099 | 0 |
| 25 30 D13S217 D13S1246 D13S171 D13S263 D13S160 D13S154 D13S173 D13S285 |
28697654 30631622 32579775 41607094 78504466 95710383 107054541 112241249 |
0.69182195 | -0.00083895 | 0 | 0.69277740 | -0.00179439 | 0 |
| 26 31 D13S1246 D13S171 D13S1694 D13S219 D13S263 D13S176 D13S154 D13S173 D13S285 |
30431300 30631622 32579775 33297510 36484263 41607094 59839530 95710383 107054541 112241249 |
0.69226137 | -0.00127836 | 0 | 0.69322965 | -0.00224664 | 0 |
| 27 32 D13S260 D13S1694 D13S263 D13S160 D13S154 |
31762621 31962968 33097286 41607094 78504466 95710383 |
0.69185808 | -0.00087507 | 0 | 0.69281315 | -0.00183014 | 0 |
| Region C | |||||||
| 28 33 D13S1695 D13S153 D13S276 D13S158 |
32849425 48416872 68335674 68536043 103224047 103424175 |
0.69148233 | -0.00049932 | 0 | 0.69242680 | -0.00144379 | 0 |
| 29 34 D13S217 D13S260 D13S1695 D13S1694 D13S219 D13S155 D13S276 D13S285 |
28697654 31962968 32849425 33297510 36484263 53695047 68335674 68536043 112041068 112241249 |
0.69142726 | -0.00044425 | 0 | 0.69237131 | -0.00138830 | 0 |
| 30 35 D13S260 D13S171 D13S267 D13S263 D13S154 |
31762621 32780083 33589982 33790250 41406784 95710383 |
0.69246020 | -0.00147720 | 0 | 0.69343471 | -0.00245171 | 0 |
| 31 36 D13S1246 D13S1695 D13S267 D13S219 D13S155 D13S176 D13S160 D13S285 |
30431300 33049658 33589982 36684592 53494781 53695047 59839530 78704740 112041068 112241249 |
0.69185880 | -0.00087579 | 0 | 0.69281373 | -0.00183073 | 0 |
| 32 37 D13S217 D13S171 D13S1695 D13S267 D13S219 D13S155 D13S153 D13S154 |
28697654 28898018 32579775 33049658 33589982 36684592 53494781 48416872 95510028 95710383 |
0.69138483 | -0.00040183 | 0 | 0.69232416 | -0.00134115 | 0 |
| 33 38 D13S260 D13S1695 D13S267 D13S155 D13S176 D13S160 D13S158 |
31762621 33049658 33589982 33790250 53494781 53695047 59839530 60039900 78504466 78704740 103224047 103424175 |
0.69137359 | -0.00039058 | 0 | 0.69231321 | -0.00133020 | 0 |
| 34 39 D13S171 D13S1695 D13S267 D13S263 D13S153 D13S276 |
32579775 33049658 33589982 41607094 48216598 48416872 68335674 68536043 |
0.69145145 | -0.00046845 | 0 | 0.69239401 | -0.00141101 | 0 |
| 35 40 D13S1694 D13S267 D13S263 D13S153 |
33097286 33790250 41406784 41607094 48216598 48416872 |
0.69135325 | -0.00037024 | 0 | 0.69229219 | -0.00130918 | 0 |
| 36 41 D13S1246 D13S1695 D13S267 D13S153 |
30431300 30631622 32849425 33049658 33589982 33790250 48216598 48416872 |
0.69135043 | -0.00036743 | 0 | 0.69228926 | -0.00130625 | 0 |
| 37 42 D13S217 D13S171 D13S267 D13S276 |
28697654 32780083 33589982 33790250 68335674 68536043 |
0.69135015 | -0.00036714 | 0 | 0.69228957 | -0.00130657 | 0 |
| 38 43 D13S1246 D13S260 D13S1695 D13S267 D13S219 D13S153 |
30431300 31962968 32849425 33049658 33589982 36684592 48216598 48416872 |
0.69138489 | -0.00040188 | 0 | 0.69232523 | -0.00134222 | 0 |
| 39 44 D13S219 D13S154 D13S173 |
36484263 36684592 95510028 107254838 |
0.69143858 | -0.00045558 | 0 | 0.69238134 | -0.00139834 | 0 |
| 40 45 D13S219 D13S153 D13S176 |
36484263 36684592 48216598 60039900 |
0.69153928 | -0.00055628 | 0 | 0.69248445 | -0.00150144 | 0 |
| Total LOH which are OK with the HGO Law (then 0) | 36 by 40 | 36 by 40 | |||||
Second analysis: 42 breast tumors LOH in Chromosome 5
Basic data: The article supporting our analysis, in the analysis from [20].
The chromosome support of our analysis: chr5:1- 181,538,259 181,538,259 bp [27].
The supporting Figure 9 of our analysis:

Figure 9: LOH pattern in breast tumors showing partial and interstitial LOH at chromosome 5 in BRCA1 mutated tumors (a), BRCA2 mutated tumors (b) and sporadic tumors (c), LOH (loss of heterozygosity); ROH (retention of heterozygosity); homozygosity or not informative. Tumors showing LOH or retention of heterozygosity at all informative markers are not shown. The chromosome regions showing highest frequencies of deletion in BRCA1 tumors according to the array CGH analysis are marked on the left side. The homozygous deletions (HZ1 and 2) detected by the array CGH analysis are indicated with arrows. Indicate an LOH target between markers D5S392 and D5S406 (4.7 Mb) in BRCA1 tumors, as well as in BRCA2 and sporadic tumors.
Figure 9 the case analysed from Figure 2 in publication [20].
To conclude: Recall that (Table 5) chromosome 5 is upstream with respect to the tipping point HGO (Figure 2) Therefore, by virtue of our "global strategy law for LOH by chromosomes", most LOHs "should" degrade the HGO value: in effect, 37 out of 42 cases degrade HGO.
Table 5 Detail of 42 breast tumors LOH deletions of chromosome 5.
| LOH references : number / ref number in published figure/Markers Id. |
Deleted LOH regions | Woman genome XX | Man genome XY | ||||
|---|---|---|---|---|---|---|---|
| Reference Human Genome (HG38 2013) | Reference HGO 0.69134779 |
Error ideal HGO – reference HGO -0.00036478 |
OK | Reference HGO 0.69228642 |
Error ideal HGO – reference HGO -0.00130341 |
OK | |
| BRCA1 Breast tumors (a) | 0 | 0 | |||||
| 3 D5S432 D5S618 D5S669 D5S404 D5S636 D5S412 |
10593074 10793430 90370500 90570824 104539362 117611769 150416827 150617121 158675514 158875859 |
0.69155669 | -0.00057368 | 0 | 0.69250257 | -0.00151957 | 0 |
| 5 D1S455 D1S398 D1S618 D1S404 D1S636 D1S412 |
35894219 58373409 90370500 117611769 150416827 150617121 158675514 158875859 |
0.69211454 | -0.00113153 | 0 | 0.69307982 | -0.00209682 | 0 |
| 6 D5S664 D5S404 D5S2011 D5S2006 |
55577693 150617121 141742521 181021853 |
0.69210365 | -0.00112064 | 0 | 0.69308176 | -0.00209875 | 0 |
| 9 D5S1977 D5S618 D5S669 D5S404 D5S636 D5S410 D5S671 D5S211 |
76918678 77119014 90370500 90570824 104539362 117611769 158675514 153495801 168195357 173921549 |
0.69147538 | -0.00049238 | 0 | 0.69241936 | -0.00143636 | 0 |
| 10 D5S664 D5S618 D5S644 D5S412 D5S671 |
55577693 55778017 90370500 96577305 158675514 168395689 |
0.69158353 | -0.00060053 | 0 | 0.69253043 | -0.00154743 | 0 |
| 13 D5S2858 D5S1977 D5S618 D5S410 D5S671 D5S2030 D5S2006 |
64525211 77119014 90370500 153495801 168195357 168395689 178280172 181021853 |
0.69189479 | -0.00091179 | 0 | 0.69285973 | -0.00187672 | 0 |
| 14 D5S664 D5S2858 D5S647 D5S620 D5S412 D5S211 D5S2006 |
55577693 55778017 64525211 67051713 82277212 158875859 141742521 141942802 180821594 181021853 |
0.69214664 | -0.00116364 | 0 | 0.69311486 | -0.00213186 | 0 |
| 17 D5S1977 D5S620 D5S2078 D5S2011 D5S2030 |
76918678 82477566 128727878 128928206 173721274 173921549 178280172 178480512 |
0.69136039 | -0.00037738 | 0 | 0.69230034 | -0.00131733 | 0 |
| 18 D5S455 D5S1977 D5S644 D5S636 D5S412 D5S211 D5S2006 |
35894219 77119014 96376990 150617121 158675514 158875859 173721274 173921549 180821594 181021853 |
0.69221927 | -0.00123627 | 0 | 0.69319501 | -0.00221201 | 0 |
| 24 D5S644 D5S669 D5S2078 |
96376990 104739650 128727878 128928206 |
0.69153474 | -0.00055174 | 0 | 0.69247922 | -0.00149622 | 0 |
| 25 D5S432 D5S455 D5S664 |
10593074 10793430 35894219 55778017 |
0.69159059 | -0.00060759 | 0 | 0.69253830 | -0.00155529 | 0 |
| 26 D5S406 D5S455 D5S398 D5S644 D5S393 D5S636 D5S671 D5S211 |
4893930 5094254 35894219 58373409 96376990 96577305 136265641 150617121 168195357 173921549 |
0.69142146 | -0.00043846 | 0 | 0.69236897 | -0.00138596 | 0 |
| BRCA2 breast tumors (b) | |||||||
| 2 D5S398 D5S636 D5S211 |
58173160 58373409 158675514 173921549 |
0.69137007 | -0.00038707 | 0 | 0.69231178 | -0.00132878 | 0 |
| 4 D5S392 D5S406 D5S618 D5S644 D5S404 D5S211 |
202025 5094254 90370500 96577305 117411476 117611769 173721274 173921549 |
0.69132495 | -0.00034195 | 1 | 0.69226494 | -0.00128194 | 1 |
| 11 D5S398 D5S647 D5S404 D5S393 D5S412 D5S671 D5S2006 |
58173160 58373409 66851322 67051713 173721274 136465819 158675514 168395689 180821594 181021853 |
0.69154086 | -0.00055785 | 0 | 0.69247962 | -0.00149661 | 0 |
| 12 D5S620 D5S618 D5S2078 |
82277212 90570824 128727878 128928206 |
0.69154344 | -0.00056043 | 0 | 0.69248811 | -0.00150511 | 0 |
| 13 D5S664 D5S647 D5S620 D5S618 |
55577693 67051713 82277212 90570824 |
0.69166055 | -0.00067754 | 0 | 0.69260989 | -0.00162689 | 0 |
| 14 D5S648 D5S644 D5S669 D5S2011 D5S636 D5S671 D5S211 |
25695625 25895922 96376990 104739650 141742521 158875859 168195357 173921549 |
0.69140625 | -0.00042325 | 0 | 0.69235149 | -0.00136848 | 0 |
| 15 D5S1997 D5S2858 D5S1977 D5S6218 D5S2078 D5S412 D5S2006 |
16882261 17082552 64525211 64725504 76918678 90570824 128727878 128928206 158675514 181021853 |
0.69137649 | -0.00039348 | 0 | 0.69232188 | -0.00133887 | 0 |
| 28 D5S392 D5S406 D5S647 D5S1977 D5S618 D5S393 D5S412 D5S671 |
202025 5094254 66851322 77119014 90370500 90570824 136265641 136465819 158675514 168395689 |
0.69137200 | -0.00038899 | 0 | 0.69231539 | -0.0013323 | 0 |
| 29 D5S392 D5S406 D5S1997 D5S664 D5S620 D5S644 D5S669 D5S2011 |
202025 5094254 16882261 17082552 55577693 82477566 96376990 104739650 141742521 141942802 |
0.69156230 | -0.00057929 | 0 | 0.69251279 | -0.00152978 | 0 |
| 31 D5S1977 D5S620 D5S644 D5S636 D5S2006 |
76918678 82477566 96376990 96577305 150416827 150617121 180821594 181021853 |
0.69136130 | -0.00037830 | 0 | 0.69230127 | -0.00131826 | 0 |
| 32 D5S2858 D5S1977 D5S620 D5S404 D5S412 D5S211 D5S2006 |
64525211 64725504 76918678 82477566 117411476 117611769 158675514 173921549 180821594 181021853 |
0.69139558 | -0.00041257 | 0 | 0.69233889 | -0.00135588 | 0 |
| 36 D5S392 D5S406 D5S636 D5S412 D5S2030 |
202025 5094254 150416827 150617121 158675514 178480512 |
0.69160321 | -0.00062020 | 0 | 0.69255168 | -0.00156867 | 0 |
| 40 D5S432 D5S455 D5S398 D5S647 |
10593074 10793430 35894219 58373409 66851322 67051713 |
0.69135641 | -0.00037340 | 0 | 0.69230022 | -0.00131721 | 0 |
| 42 D5S620 D5S644 D5S404 D5S2011 D5S671 D5S2030 D5S2006 |
82277212 82477566 96376990 96577305 117411476 117611769 141742521 168395689 178280172 181021853 |
0.69102838 | -0.00004537 | 1 | 0.69196161 | -0.00097860 | 1 |
| Sporadic (c) | |||||||
| 5 D5S406 D5S398 D5S1977 D5S2011 D5S410 |
4893930 5094254 58173160 77119014 141742521 153495801 |
0.69148573 | -0.00050272 | 0 | 0.69243277 | -0.00144976 | 0 |
| 6 D5S648 D5S664 D5S398 D5S1977 D5S2011 |
25695625 25895922 55577693 58373409 76918678 77119014 141742521 141942802 |
0.69135535 | -0.00037235 | 0 | 0.69229472 | -0.00131172 | 0 |
| 9 D5S644 D5S404 D5S636 D5S412 |
96376990 117611769 150416827 150617121 158675514 158875859 |
0.69173687 | -0.00075387 | 0 | 0.69268834 | -0.00170534 | 0 |
| 10 D5S620 D5S644 D5S2078 D5S412 D5S211 D5S2006 |
82277212 82477566 96376990 96577305 128727878 158875859 173721274 173921549 180821594 181021853 |
0.69121921 | -0.00023620 | 1 | 0.69215990 | -0.00117690 | 1 |
| 11 D5S406 D5S432 D5S455 |
4893930 10793430 35894219 36094418 |
0.69132699 | -0.00034399 | 1 | 0.69226614 | -0.00128313 | 1 |
| 12 D5S432 D5S648 D5S647 D5S620 D5S2078 D5S2011 D5S410 |
10593074 10793430 25695625 67051713 82277212 128928206 173721274 153495801 |
0.69275045 | -0.00176744 | 0 | 0.69373407 | -0.00275107 | 0 |
| 13 D5S398 D5S647 D5S644 D5S393 D5S412 D5S211 |
58173160 58373409 66851322 67051713 96376990 96577305 136265641 158875859 173721274 173921549 |
0.69125826 | -0.00027525 | 1 | 0.69219863 | -0.00121563 | 1 |
| 15 D5S455 D5S2858 D5S2006 |
35894219 64725504 180821594 181021853 |
0.69168749 | -0.00070448 | 0 | 0.69263900 | -0.00165600 | 0 |
| 19 D5S392 D5S455 D5S398 D5S2858 D5S644 D5S669 D5S410 D5S2030 |
202025 36094418 58173160 64725504 96376990 104739650 153295415 153495801 178280172 178480512 |
0.69186327 | -0.00088027 | 0 | 0.69282275 | -0.00183974 | 0 |
| 21 D5S392 D5S398 D5S647 D5S404 D5S2078 D5S636 D5S410 |
202025 402151 58173160 58373409 66851322 67051713 117411476 128928206 150416827 153495801 |
0.69151381 | -0.00053081 | 0 | 0.6924589 | -0.00147589 | 0 |
| 24 D5S664 D5S620 D5S644 D5S404 |
55577693 82477566 96376990 96577305 117411476 117611769 |
0.69154041 | -0.00055740 | 0 | 0.69248815 | -0.00150515 | 0 |
| 26 D5S664 D5S398 D5S2078 D5S636 |
55577693 58373409 128727878 128928206 150416827 150617121 |
0.69135403 | -0.00037102 | 0 | 0.69229333 | -0.00131033 | 0 |
| 31 D5S636 D5S412 D5S671 |
150416827 150617121 158675514 168395689 |
0.69145662 | -0.00047361 | 0 | 0.69239947 | -0.00141646 | 0 |
| 33 D5S648 D5S2858 D5S618 D5S644 D5S2078 D5S211 |
25695625 25895922 64525211 64725504 90370500 96577305 128727878 128928206 173721274 173921549 |
0.69147158 | -0.00048858 | 0 | 0.69241431 | -0.00143130 | 0 |
| 34 D5S2858 D5S647 D5S620 D5S618 D5S404 D5S2011 D5S410 D5S671 |
64525211 67051713 82277212 90570824 117411476 117611769 141742521 153495801 168195357 168395689 |
0.69155438 | -0.00057137 | 0 | 0.69250173 | -0.00151872 | 0 |
| 36 D5S455 D5S398 D5S2078 D5S2011 |
35894219 58373409 128727878 128928206 141742521 141942802 |
0.69160183 | -0.00061883 | 0 | 0.69255027 | -0.00156727 | 0 |
| Total LOH which are OK with the HGO Law (then 0) | 37 by 42 | 37 by 42 | |||||
Third analysis: 72 prostate tumors LOH in chromosome 16
Basic data: The article supporting our analysis [24].
The chromosome support of our analysis: chr16:1- 90,338,345 90,338,345 bp [28].
The supporting Figure 10 of our analysis:
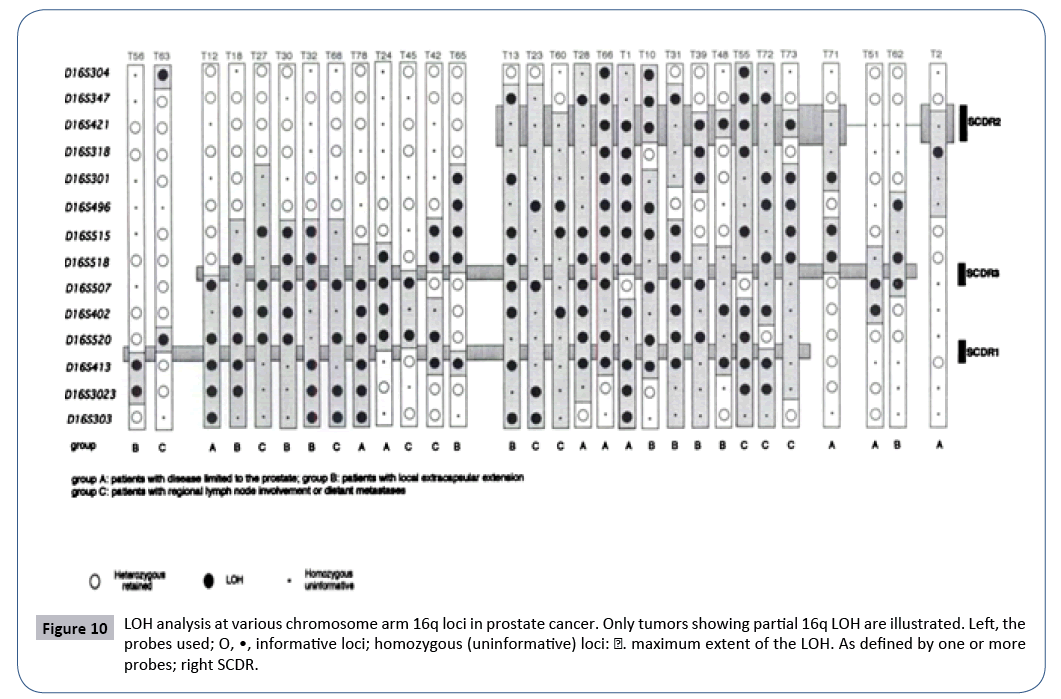
Figure 10: LOH analysis at various chromosome arm 16q loci in prostate cancer. Only tumors showing partial 16q LOH are illustrated. Left, the probes used; O, •, informative loci; homozygous (uninformative) loci: ∎. maximum extent of the LOH. As defined by one or more probes; right SCDR.
Figure 10 the case analysed from Figure 1 in publication [24].
To conclude: Recall that (Table 6) chromosome 16 is downstream with respect to the tipping point HGO (Figure 3). Therefore, by virtue of our "global LOH strategy law by chromosomes", most LOHs "should" improve the HGO value: in fact, 66 cases out of 72 increase the HGO in the case of a Genome (XX) but 71 out of 72 cases increase HGO in the case of a male genome (XY).
Table 6 Detail of 72 prostate tumors LOH deletions of chromosome 16.
| LOH references : number / ref number in published figure/Markers Id. |
Deleted LOH regions | Woman genome XX | Man genome XY | ||||
|---|---|---|---|---|---|---|---|
| Reference Human Genome (HG38 2013) | Reference HGO 0.69134779 |
Error ideal HGO – reference HGO -0.00036478 |
OK | Reference HGO 0.69228642 |
Error ideal HGO – reference HGO -0.00130341 |
OK | |
| Regions 1/3 Minimal |
|||||||
| 1 T56 D16S413 D16S3023 |
87760230 88555205 | 0.69129368 | -0.00031068 | 1 | 0.69223116 | -0.00124815 | 1 |
| 2 T63 D16S304 D16S520 |
86382506 86582729 | 0.69133837 | -0.00035536 | 1 | 0.69227681 | -0.00129380 | 1 |
| 3 T12 D16S507 D16S303 |
79942534 90182695 | 0.69094866 | 0.00003434 | 1 | 0.69187951 | -0.00089651 | 1 |
| 4 T18 D16S518 D16S3023 |
78004124 88555205 | 0.69104347 | -0.00006046 | 1 | 0.69197662 | -0.00099361 | 1 |
| 5 T27 D16S515 D16S520 |
76383131 86582729 | 0.69115928 | -0.00017628 | 1 | 0.69209512 | -0.00111212 | 1 |
| 6 T30 D16S515 D16S520 |
76383131 86582729 | 0.69115928 | -0.00017628 | 1 | 0.69209512 | -0.00111212 | 1 |
| 7 T32 D16S515 D16S303 |
76383131 90182695 | 0.69094149 | 0.00004152 | 1 | 0.69187275 | -0.00088975 | 1 |
| 8 T68 D16S507 D16S303 |
79942534 90182695 | 0.69094866 | 0.00003434 | 1 | 0.69187951 | -0.00089651 | 1 |
| 9 T78 D16S507 D16S303 |
79942534 90182695 | 0.69094866 | 0.00003434 | 1 | 0.69187951 | -0.00089651 | 1 |
| 10 T24 D16S518 D16S520 |
78004124 86582729 | 0.69114909 | -0.00016609 | 1 | 0.69208443 | -0.00110142 | 1 |
| 11 T45 D16S507 D16S520 |
79942534 86582729 | 0.69116632 | -0.00018332 | 1 | 0.69210174 | -0.00111874 | 1 |
| 12 T42 D16S515 D16S413 |
76383131 87960566 | 0.69109574 | -0.00011274 | 1 | 0.69203030 | -0.00104730 | 1 |
| 13 T65 D16S301 D16S413 |
66802038 87960566 | 0.69092211 | 0.00006089 | 1 | 0.69185412 | -0.00087111 | 1 |
| 14 T13 D16S347 D16S303 |
7161888 90182695 | 0.69019295 | 0.00079006 | 0 | 0.69111633 | -0.00013333 | 1 |
| 15 T23 D16S496 D16S303 |
68814802 90182695 | 0.69085034 | 0.00013267 | 1 | 0.69178068 | -0.00079767 | 1 |
| 16 T60 D16S496 D16S402 |
68814802 83360293 | 0.69120855 | -0.00022555 | 1 | 0.69214628 | -0.00116328 | 1 |
| 17 T28 D16S347 D16S413 |
7161888 87960566 | 0.69034898 | 0.00063402 | 0 | 0.69127574 | -0.00029274 | 1 |
| 18 T66 D16S304 D16S413 |
7161888 87960566 | 0.69034898 | 0.00063402 | 0 | 0.69127574 | -0.00029274 | 1 |
| 19 T1 D16S421 D16S303 |
67161726 90182695 | 0.69078070 | 0.00020231 | 1 | 0.69170965 | -0.00072664 | 1 |
| 20 T10 D16S304 D16S413 |
7161888 87960566 | 0.69034898 | 0.00063402 | 0 | 0.69127574 | -0.00029274 | 1 |
| 21 T31 D16S304 D16S413 |
7161888 87960566 | 0.69034898 | 0.00063402 | 0 | 0.69127574 | -0.00029274 | 1 |
| 22 T39 D16S421 D16S520 |
67161726 86582729 | 0.69099890 | -0.00001589 | 1 | 0.69193244 | -0.00094944 | 1 |
| 23 T48 D16S421 D16S413 |
67161726 87960566 | 0.69093523 | 0.00004778 | 1 | 0.69186749 | -0.00088448 | 1 |
| 24 T55 D16S402 D16S3023 |
67161726 88555205 | 0.69089304 | 0.00008997 | 1 | 0.69182440 | -0.00084139 | 1 |
| 25 T72 D16S402 D16S3023 |
67161726 88555205 | 0.69089304 | 0.00008997 | 1 | 0.69182440 | -0.00084139 | 1 |
| 26 T73 D16S421 D16S518 |
67161726 78204435 | 0.69119627 | -0.00021327 | 1 | 0.69213313 | -0.00115013 | 1 |
| 27 T71 D16S301 D16S518 |
66802038 78204435 | 0.69118319 | -0.00020019 | 1 | 0.69211980 | -0.00113679 | 1 |
| 28 T51 D16S507 D16S402 |
79942534 83360293 | 0.69130612 | -0.00032311 | 1 | 0.69224433 | -0.00126132 | 1 |
| 29 T62 D16S496 D16S507 |
68814802 80142885 | 0.69125013 | -0.00026713 | 1 | 0.69218832 | -0.00120531 | 1 |
| 30 T2 D16S318 |
67161888 67362019 | 0.69133725 | -0.00035425 | 1 | 0.69227567 | -0.00129266 | 1 |
| 2/3 Regions | |||||||
| 31 T12 D16S507 D16S520 D16S303 |
79942534 80142885 86382506 90182695 |
0.69112078 | -0.00013778 | 1 | 0.69205469 | -0.00107169 | 1 |
| 32 T18 D16S518 D16S402 D16S3023 |
78004124 78204435 83159809 88555205 |
0.69110000 | -0.00011699 | 1 | 0.69203368 | -0.00105067 | 1 |
| 33 T27 D16S515 D16S507 D16S520 |
76383131 76583505 79942534 86582729 |
0.69117068 | -0.00018767 | 1 | 0.69210623 | -0.00112323 | 1 |
| 34 T32 D16S515 D16S507 D16S413 D16S303 |
76383131 80142885 87760230 90182695 |
0.69117454 | -0.00019154 | 1 | 0.69211008 | -0.00112708 | 1 |
| 35 T68 D16S507 D16S520 D16S3023 D16S303 |
79942534 80142885 86382506 86582729 88354919 90182695 |
0.69120697 | -0.00022396 | 1 | 0.69214262 | -0.00115962 | 1 |
| 36 T45 D16S507 D16S520 |
79942534 80142885 86382506 86582729 |
0.69133815 | -0.00035514 | 1 | 0.69227661 | -0.00129361 | 1 |
| 37 T42 D16S515 D16S518 D16S520 D16S413 |
76383131 78204435 86382506 87960566 |
0.69128331 | -0.00030030 | 1 | 0.69222097 | -0.00123796 | 1 |
| 38 T65 D16S301 D16S518 D16S413 |
66802038 78204435 87760230 87960566 |
0.69117107 | -0.00018806 | 1 | 0.69210742 | -0.00112442 | 1 |
| 39 T13 D16S421 D16S301 D16S515 D16S402 D16S413 D16S303 |
67161726 67362083 66802038 67002181 76383131 83360293 87760230 87960566 89982590 90182695 |
0.69128322 | -0.00030022 | 1 | 0.69222151 | -0.00123851 | 1 |
| 40 T23 D16S 496 D16S507 D16S3023 D16S303 |
68814802 69015161 79942534 80142885 88354919 90182695 |
0.69121462 | -0.00023162 | 1 | 0.69215046 | -0.00116746 | 1 |
| 41 T60 D16S496 D16S515 D16S402 |
68814802 76583505 83159809 83360293 |
0.69126101 | -0.00027800 | 1 | 0.69219890 | -0.00121589 | 1 |
| 42 T28 D16S347 D16S515 D16S413 |
67161726 67362083 76383131 87960566 |
0.69108516 | -0.00010216 | 1 | 0.69201950 | -0.00103650 | 1 |
| 43 T66 D16S304 D16S507 D16S520 D16S413 |
67161726 80142885 86382506 87960566 |
0.69110767 | -0.00012467 | 1 | 0.69204301 | -0.00106000 | 1 |
| 44 T1 D16S421 D16S518 D16S402 D16S413 D16S303 |
67161726 78204435 83159809 83360293 87760230 90182695 |
0.69102923 | -0.00004622 | 1 | 0.69196255 | -0.00097954 | 1 |
| 45 T10 D16S304 D16S496 D16S515 D16S507 D16S520 D16S413 |
67161726 67362083 68814802 76583505 79942534 80142885 86382506 87960566 |
0.69117796 | -0.00019496 | 1 | 0.69211417 | -0.00113117 | 1 |
| 46 T31 D16S347 D16S515 D16S413 |
67161726 67362083 76383131 87960566 |
0.69108516 | -0.00010216 | 1 | 0.69201950 | -0.00103650 | 1 |
| 47 T39 D16S421 D16S301 D16S507 D16S520 |
67161726 67002181 79942534 86582729 |
0.69117039 | -0.00018738 | 1 | 0.69210588 | -0.00112287 | 1 |
| 48 T48 D16S421 D16S402 D16S413 |
67161726 67362083 83159809 87960566 |
0.69113337 | -0.00015037 | 1 | 0.69206774 | -0.00108474 | 1 |
| 49 T55 D16S304 D16S318 D16S515 D16S402 D16S3023 |
67161726 67362019 76383131 76583505 83159809 88555205 |
0.69109567 | -0.00011266 | 1 | 0.69202927 | -0.00104627 | 1 |
| 50 T72 D16S347 D16S421 D16S301 D16S496 D16S518 D16S402 D16S413 D16S3023 |
67161726 67362083 66802038 69015161 78004124 78204435 83159809 83360293 87760230 88555205 |
0.69122003 | -0.00023703 | 1 | 0.69215614 | -0.00117313 | 1 |
| 51 T73 D16S421 D16S301 D16S518 |
67161726 67362083 66802038 78204435 |
0.69119627 | -0.00021327 | 1 | 0.69213313 | -0.00115013 | 1 |
| 52 T71 D16S301 D16S515 D16S518 |
66802038 67002181 76383131 78204435 |
0.69134716 | -0.00036415 | 1 | 0.69228611 | -0.00130310 | 1 |
| 53 T62 D16S496 D16S518 D16S507 |
68814802 69015161 78004124 80142885 |
0.69132865 | -0.00034564 | 1 | 0.69226721 | -0.00128421 | 1 |
| Regions 3/3 Maximal |
1 | 1 | |||||
| 54 T18 D16S515 D16S303 |
76383131 90182695 | 0.69094149 | 0.00004152 | 1 | 0.69187275 | -0.00088975 | 1 |
| 55 T27 D16S301 D16S303 |
66802038 90182695 | 0.69076757 | 0.00021544 | 1 | 0.69169627 | -0.00071326 | 1 |
| 56 T78 D16S518 D16S303 |
78004124 90182695 | 0.69093136 | 0.00005165 | 1 | 0.69186211 | -0.00087911 | 1 |
| 57 T23 D16S347 D16S303 |
67161726 90182695 | 0.69078070 | 0.00020231 | 1 | 0.69170965 | -0.00072664 | 1 |
| 58 T28 D16S421 D16S3023 |
67161726 88555205 | 0.69089304 | 0.00008997 | 1 | 0.69182440 | -0.00084139 | 1 |
| 59 T66 D16S421 D16S413 |
67161726 87960566 | 0.69093523 | 0.00004778 | 1 | 0.69186749 | -0.00088448 | 1 |
| 60 T71 D16S421 D16S518 |
67161726 78204435 | 0.69119627 | -0.00021327 | 1 | 0.69213313 | -0.00115013 | 1 |
| Autres multiples maximums | 1 | 1 | |||||
| 61 T42 D16S515 D16S518 D16S402 D16S413 |
76383131 78204435 83159809 87960566 |
0.69115224 | -0.00016924 | 1 | 0.69208733 | -0.00110432 | 1 |
| 62 T10 D16S421 D16S301 D16S413 |
67161726 67362083 66802038 87960566 |
0.69093523 | 0.00004778 | 1 | 0.69186749 | -0.00088448 | 1 |
| 63 T31 D16S421 D16S301 D16S515 D16S303 |
67161726 67002181 76383131 90182695 |
0.69094556 | 0.00003744 | 1 | 0.69187690 | -0.00089389 | 1 |
| 64 T39 D16S421 D16S301 D16S518 D16S303 |
67161726 67002181 78004124 90182695 |
0.69093543 | 0.00004757 | 1 | 0.69186626 | -0.00088325 | 1 |
| 65 T48 D16S421 D16S518 D16S413 |
67161726 67362083 78004124 87960566 |
0.69107499 | -0.00009199 | 1 | 0.69200883 | -0.00102582 | 1 |
| 66 T1 D16S421 D16S518 D16S402 D16S303 |
67161726 78204435 83159809 90182695 |
0.69083756 | 0.00014545 | 1 | 0.69176705 | -0.00078404 | 1 |
| 67 T10 D16S421 D16S301 D16S413 |
67161726 67362083 66802038 87960566 |
0.69093523 | 0.00004778 | 1 | 0.69186749 | -0.00088448 | 1 |
| 68 T72 D16S421 D16S402 D16S413 D16S303 |
67161726 83360293 87760230 90182695 |
0.69097256 | 0.00001045 | 1 | 0.69190535 | -0.00092234 | 1 |
| 69 T73 D16S421 D16S301 D16S3023 |
67161726 67362083 66802038 88555205 |
0.69089304 | 0.00008997 | 1 | 0.69182440 | -0.00084139 | 1 |
| 70 T71 D16S421 D16S301 D16S515 D16S518 |
67002181 67161726 76383131 78204435 |
0.69135616 | -0.00037315 | 0 | 0.69229529 | -0.00131228 | 0 |
| 71 T51 D16S518 D16S402 |
78004124 83360293 | 0.69128895 | -0.00030594 | 1 | 0.69222706 | -0.00124406 | 1 |
| 72 T2 D16S421 D16S496 |
67161726 69015161 | 0.69127674 | -0.00029373 | 1 | 0.69221399 | -0.00123098 | 1 |
| Total LOH which are OK with the HGO Law (then 1) | 66 by 72 | 71 by 72 | |||||
Discussion
First analysis: 40 breast tumors LOH in chromosome 13
In the analysis from publication [19], we show that HGO conforms to the above law in 36 of 40 cases (chromosome 13 is in the region upstream of the HGO point) (Figure 2).
In Figure 11, it is interesting to note that the effect on a female genome (XX) is greater than the effect on this same male genome (XY), which is in agreement with the fact that this cancer is almost exclusively feminine. A rapid analysis of the degradations of HGO - for the same LOH - depending on whether the genome is XY (masculine) or XX (feminine) will show that these expected degradations are 1.27 times higher in the case of a female genome (XX). This would reinforce the fact that the female genome is more sensitive to these LOH deletions than the male genome.
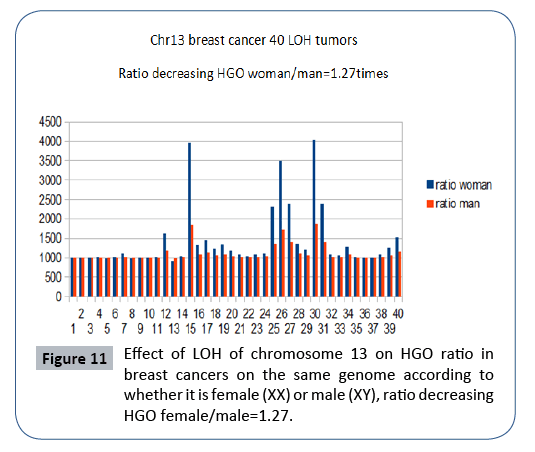
Figure 11: Effect of LOH of chromosome 13 on HGO ratio in breast cancers on the same genome according to whether it is female (XX) or male (XY), ratio decreasing HGO female/male=1.27.
Second analysis: 42 breast tumors LOH in chromosome 5
In the analysis from publication [20], we show (Figure 12) that the HGO conforms to the above-mentioned law in 37 cases out of 42 (chromosome 5 is located in the region upstream of the HGO point) (Figure 2). The analysis below concerns cases of LOH affecting BRCA1 mutations. It is interesting to observe - even if only visually - that the effect on a female genome (XX) is greater than the effect on the same male genome (XY), which is in agreement with the fact that this cancer is almost exclusively feminine.
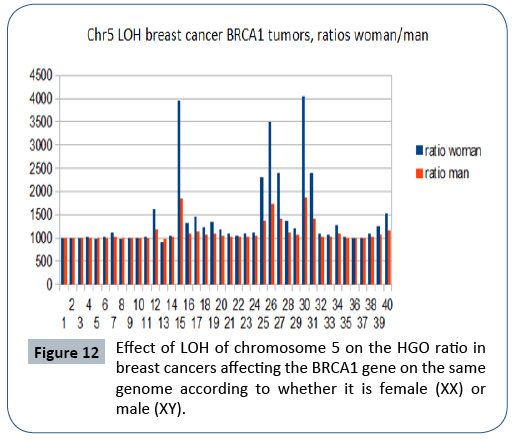
Figure 12: Effect of LOH of chromosome 5 on the HGO ratio in breast cancers affecting the BRCA1 gene on the same genome according to whether it is female (XX) or male (XY).
Third analysis: 72 prostate tumors LOH in chromosome 16
Of 72 cases analyzed from article [24], 71 cases improved HGO while only one did not (Chromosome 16 is located in the region downstream of the HGO point (Figure 3). The evaluation below analyzes the evolution of the HGO according to the 3 grades of pathogenicity of the tumors studied.
Grade A: patients with disease limited to the prostate.
Grade B: patients with local extracapsular extension.
Grade C: patients with regional lymph node involvement or distant metastases.
Between Grade A and Grade B, an average HGO improvement of 4.88% was observed.
The graph below (Figure 13) illustrates this optimization. The HGO of grade C is of the same order as that of grade A, which may be explained by the fact that the LOHs of grades A and B are generally monochromosomes whereas the LOHs of grade C are multichromosomes. There are therefore more complexes to be demonstrated in monochromosome studies.
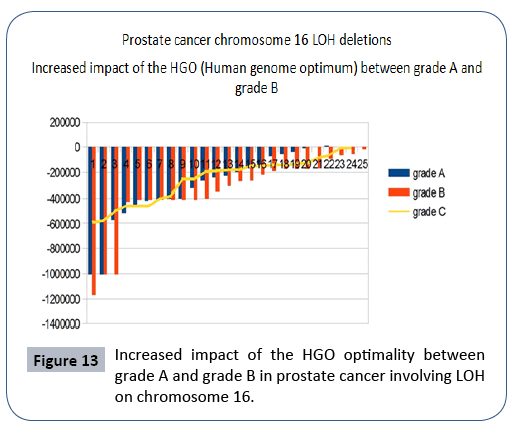
Figure 13: Increased impact of the HGO optimality between grade A and grade B in prostate cancer involving LOH on chromosome 16.
It is interesting to note that the effect on a female genome (XX) is 4.86 times lower than the effect on the same male genome (XY), which is in agreement with the fact that this cancer is exclusively male (Figure 14).
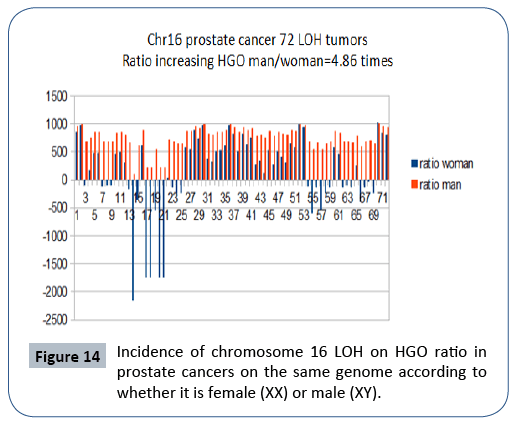
Figure 14: Incidence of chromosome 16 LOH on HGO ratio in prostate cancers on the same genome according to whether it is female (XX) or male (XY).
Conclusion
It is remarkable to discover this strong correlation between the pathogenicity and the aggressiveness of tumors on the one hand, and the categorization and hierarchization of mutations LOH of the mutant genomes at the origin of tumors. The law discovered here is universal, common to all types of cancers and to all chromosomes. Its nature is mathematical, proceeding only from the DNA information of the genomes. The method is exhaustive, systematic, and predictive, making it possible to anticipate the knowledge of the pathogenicity of an existing tumor or even of a mutation before the tumor disappears.
The mutant genome thus constitutes a kind of signature, virtual and numerical image of the tumor. Thus it is remarkable to discover that the LOH genome of a breast tumor, a cancer that is exclusively feminine by its nature, will lead to an overall HGO genomic degradation much greater than the same mutation affecting a male genome.
Conversely, it will be the same, but in the opposite way for a cancer of the prostate which, it is, exclusively masculine.
Finally, in this same prostate cancer, the impact on HGO between grade A and grade B is significant: recall.
Grade A: patients with disease limited to the prostate.
Grade B: patients with local extracapsular extension.
Grade C: patients with regional lymph node involvement or distant metastases.
Between Grade A and Grade B, an average HGO improvement of 4.88% was observed.
To conclude, the most important fact is the universal character of this HGO law, presented here for both breast and prostate cancers, the universal character of this law has been widely generalized experimentally on a large number of cancers differences affecting most chromosomes, this generalization will be described in [28,29].
Acknowledgement
We especially thank Dr. Robert Friedman M.D. practiced nutritional and preventive medicine in Santa Fe, New Mexico, woldwide expert on Golden ratio Life applications. We also thank the mathematician Pr. Diego Lucio Rapoport (Buenos aires), the french biologist Pr. François Gros (Pasteur institute, co-discoverer of RNA messenger with James Watson and Walter Gilbert) and Pr. Luc Montagnier, medicine Nobel prizewinner for their interest in my research of biomathematical laws of genomes.